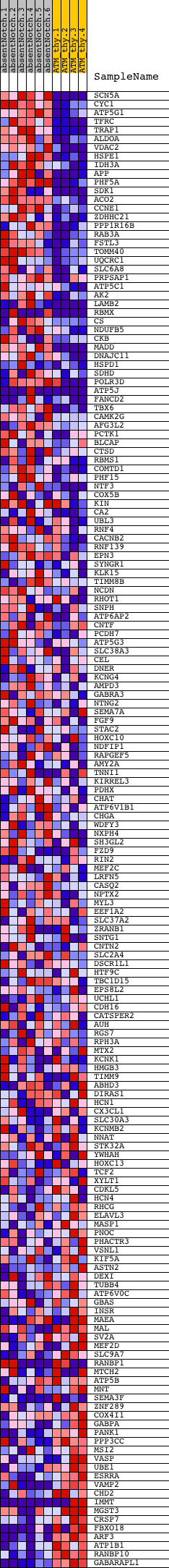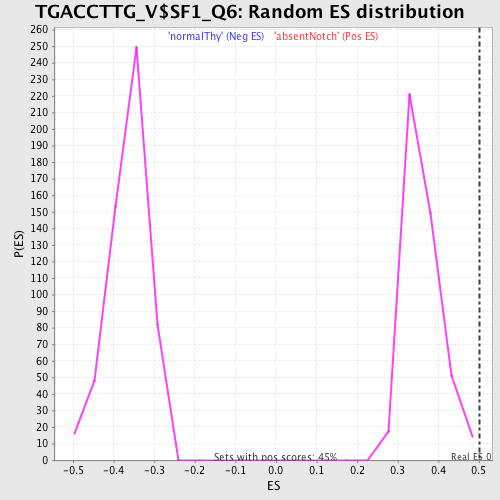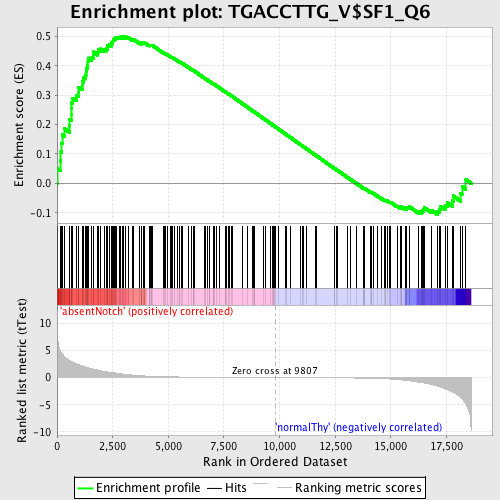
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | TGACCTTG_V$SF1_Q6 |
| Enrichment Score (ES) | 0.50114805 |
| Normalized Enrichment Score (NES) | 1.3901323 |
| Nominal p-value | 0.0044247787 |
| FDR q-value | 0.43666032 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SCN5A | 4070017 | 27 | 7.351 | 0.0496 | Yes | ||
| 2 | CYC1 | 2850546 | 154 | 4.832 | 0.0763 | Yes | ||
| 3 | ATP5G1 | 5290019 | 171 | 4.688 | 0.1080 | Yes | ||
| 4 | TFRC | 4050551 | 213 | 4.421 | 0.1364 | Yes | ||
| 5 | TRAP1 | 6040168 | 228 | 4.327 | 0.1657 | Yes | ||
| 6 | ALDOA | 6290672 | 348 | 3.761 | 0.1854 | Yes | ||
| 7 | VDAC2 | 7000215 | 560 | 3.129 | 0.1957 | Yes | ||
| 8 | HSPE1 | 3170438 | 569 | 3.119 | 0.2169 | Yes | ||
| 9 | IDH3A | 460332 | 630 | 2.963 | 0.2342 | Yes | ||
| 10 | APP | 2510053 | 634 | 2.946 | 0.2545 | Yes | ||
| 11 | PHF5A | 2690519 | 652 | 2.899 | 0.2737 | Yes | ||
| 12 | SDK1 | 2850139 | 705 | 2.812 | 0.2904 | Yes | ||
| 13 | ACO2 | 4230600 | 888 | 2.467 | 0.2977 | Yes | ||
| 14 | CCNE1 | 110064 | 949 | 2.374 | 0.3109 | Yes | ||
| 15 | ZDHHC21 | 2370338 | 957 | 2.357 | 0.3269 | Yes | ||
| 16 | PPP1R16B | 3520300 | 1125 | 2.158 | 0.3328 | Yes | ||
| 17 | RAB3A | 3940288 | 1130 | 2.154 | 0.3475 | Yes | ||
| 18 | FSTL3 | 5420279 | 1188 | 2.074 | 0.3588 | Yes | ||
| 19 | TOMM40 | 6110017 | 1256 | 1.984 | 0.3690 | Yes | ||
| 20 | UQCRC1 | 520053 | 1300 | 1.940 | 0.3801 | Yes | ||
| 21 | SLC6A8 | 3940341 | 1316 | 1.924 | 0.3927 | Yes | ||
| 22 | PRPSAP1 | 6980494 | 1366 | 1.853 | 0.4029 | Yes | ||
| 23 | ATP5C1 | 5910601 | 1387 | 1.828 | 0.4145 | Yes | ||
| 24 | AK2 | 4590458 4760195 6110097 | 1395 | 1.814 | 0.4267 | Yes | ||
| 25 | LAMB2 | 4920184 | 1565 | 1.609 | 0.4287 | Yes | ||
| 26 | RBMX | 7100162 2900541 | 1619 | 1.566 | 0.4367 | Yes | ||
| 27 | CS | 5080600 | 1623 | 1.562 | 0.4474 | Yes | ||
| 28 | NDUFB5 | 110685 | 1823 | 1.382 | 0.4462 | Yes | ||
| 29 | CKB | 4560632 | 1843 | 1.370 | 0.4547 | Yes | ||
| 30 | MADD | 6100725 7000088 | 1954 | 1.261 | 0.4574 | Yes | ||
| 31 | DNAJC11 | 360465 3390215 | 2110 | 1.132 | 0.4569 | Yes | ||
| 32 | HSPD1 | 60097 2030451 | 2231 | 1.033 | 0.4576 | Yes | ||
| 33 | SDHD | 2570128 | 2260 | 1.023 | 0.4632 | Yes | ||
| 34 | POLR3D | 580037 6420615 | 2262 | 1.022 | 0.4702 | Yes | ||
| 35 | ATP5J | 4210100 6900288 | 2343 | 0.997 | 0.4728 | Yes | ||
| 36 | FANCD2 | 1850280 6840348 | 2448 | 0.942 | 0.4737 | Yes | ||
| 37 | TBX6 | 4730139 6840647 | 2460 | 0.935 | 0.4796 | Yes | ||
| 38 | CAMK2G | 630097 | 2508 | 0.898 | 0.4833 | Yes | ||
| 39 | AFG3L2 | 5130332 | 2512 | 0.897 | 0.4893 | Yes | ||
| 40 | PCTK1 | 4560079 | 2576 | 0.862 | 0.4919 | Yes | ||
| 41 | BLCAP | 3990253 | 2620 | 0.829 | 0.4953 | Yes | ||
| 42 | CTSD | 1070195 | 2682 | 0.791 | 0.4975 | Yes | ||
| 43 | RBMS1 | 6400014 | 2796 | 0.721 | 0.4964 | Yes | ||
| 44 | COMTD1 | 1170193 | 2851 | 0.695 | 0.4983 | Yes | ||
| 45 | PHF15 | 6550066 6860673 | 2930 | 0.658 | 0.4986 | Yes | ||
| 46 | NTF3 | 5890131 | 2966 | 0.636 | 0.5011 | Yes | ||
| 47 | COX5B | 6550670 | 3093 | 0.576 | 0.4983 | No | ||
| 48 | KIN | 6550014 | 3209 | 0.525 | 0.4957 | No | ||
| 49 | CA2 | 1660113 1660600 | 3383 | 0.454 | 0.4895 | No | ||
| 50 | UBL3 | 6450458 | 3429 | 0.440 | 0.4901 | No | ||
| 51 | RNF4 | 580286 1400152 | 3681 | 0.358 | 0.4790 | No | ||
| 52 | CACNB2 | 1500095 7330707 | 3778 | 0.328 | 0.4761 | No | ||
| 53 | RNF139 | 2650114 3610450 | 3783 | 0.327 | 0.4781 | No | ||
| 54 | EPN3 | 2350592 | 3808 | 0.321 | 0.4790 | No | ||
| 55 | SYNGR1 | 5130452 6650056 | 3875 | 0.304 | 0.4776 | No | ||
| 56 | KLK15 | 6350086 | 3923 | 0.293 | 0.4771 | No | ||
| 57 | TIMM8B | 1190364 | 3945 | 0.288 | 0.4779 | No | ||
| 58 | NCDN | 50040 1980739 3520603 | 4138 | 0.248 | 0.4692 | No | ||
| 59 | RHOT1 | 6760692 | 4145 | 0.246 | 0.4706 | No | ||
| 60 | SNPH | 2680484 | 4187 | 0.239 | 0.4701 | No | ||
| 61 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 4227 | 0.232 | 0.4696 | No | ||
| 62 | CNTF | 450128 870253 1780068 6520195 | 4243 | 0.231 | 0.4703 | No | ||
| 63 | PCDH7 | 450398 4150040 | 4265 | 0.227 | 0.4708 | No | ||
| 64 | ATP5G3 | 4230551 | 4782 | 0.156 | 0.4439 | No | ||
| 65 | SLC38A3 | 5340142 | 4831 | 0.151 | 0.4423 | No | ||
| 66 | CEL | 2640278 | 4878 | 0.147 | 0.4409 | No | ||
| 67 | DNER | 3610170 | 4974 | 0.138 | 0.4367 | No | ||
| 68 | KCNG4 | 1850736 5050113 | 5106 | 0.128 | 0.4305 | No | ||
| 69 | AMPD3 | 610369 3360528 | 5132 | 0.126 | 0.4300 | No | ||
| 70 | GABRA3 | 4810142 | 5192 | 0.120 | 0.4276 | No | ||
| 71 | NTNG2 | 1770438 6380408 | 5262 | 0.115 | 0.4247 | No | ||
| 72 | SEMA7A | 2630239 5050593 | 5414 | 0.106 | 0.4172 | No | ||
| 73 | FGF9 | 1050195 | 5498 | 0.101 | 0.4134 | No | ||
| 74 | STAC2 | 7100022 | 5515 | 0.100 | 0.4133 | No | ||
| 75 | HOXC10 | 5360170 | 5573 | 0.097 | 0.4108 | No | ||
| 76 | NDFIP1 | 1980402 | 5641 | 0.094 | 0.4079 | No | ||
| 77 | RAPGEF5 | 1500603 3450427 6130056 | 5897 | 0.083 | 0.3946 | No | ||
| 78 | AMY2A | 580138 | 6049 | 0.077 | 0.3870 | No | ||
| 79 | TNNI1 | 3710040 | 6113 | 0.075 | 0.3841 | No | ||
| 80 | KIRREL3 | 2630195 | 6140 | 0.074 | 0.3832 | No | ||
| 81 | PDHX | 870315 | 6154 | 0.073 | 0.3830 | No | ||
| 82 | CHAT | 6840603 | 6633 | 0.058 | 0.3575 | No | ||
| 83 | ATP6V1B1 | 6100181 | 6673 | 0.057 | 0.3557 | No | ||
| 84 | CHGA | 1990056 6550463 | 6748 | 0.054 | 0.3521 | No | ||
| 85 | WDFY3 | 3610041 4560333 4570273 | 6835 | 0.052 | 0.3478 | No | ||
| 86 | NXPH4 | 3830500 | 6853 | 0.051 | 0.3472 | No | ||
| 87 | SH3GL2 | 1990176 3830494 | 7031 | 0.047 | 0.3380 | No | ||
| 88 | FZD9 | 5360136 | 7042 | 0.047 | 0.3378 | No | ||
| 89 | RIN2 | 2450184 | 7078 | 0.046 | 0.3362 | No | ||
| 90 | MEF2C | 670025 780338 | 7167 | 0.044 | 0.3317 | No | ||
| 91 | LRFN5 | 6650088 | 7303 | 0.042 | 0.3247 | No | ||
| 92 | CASQ2 | 3170136 | 7556 | 0.037 | 0.3113 | No | ||
| 93 | NPTX2 | 70021 | 7576 | 0.036 | 0.3105 | No | ||
| 94 | MYL3 | 6040563 | 7617 | 0.035 | 0.3086 | No | ||
| 95 | EEF1A2 | 2260162 5290086 | 7714 | 0.033 | 0.3036 | No | ||
| 96 | SLC37A2 | 2970750 | 7737 | 0.033 | 0.3026 | No | ||
| 97 | ZRANB1 | 510373 4540278 | 7834 | 0.031 | 0.2977 | No | ||
| 98 | SNTG1 | 6770373 | 7866 | 0.031 | 0.2962 | No | ||
| 99 | CNTN2 | 2760204 | 8338 | 0.023 | 0.2708 | No | ||
| 100 | SLC2A4 | 540441 | 8543 | 0.020 | 0.2599 | No | ||
| 101 | DSCR1L1 | 940438 | 8801 | 0.015 | 0.2461 | No | ||
| 102 | HTF9C | 3190048 4120132 | 8805 | 0.015 | 0.2460 | No | ||
| 103 | TBC1D15 | 870673 | 8853 | 0.014 | 0.2436 | No | ||
| 104 | EPS8L2 | 2850528 | 8885 | 0.014 | 0.2420 | No | ||
| 105 | UCHL1 | 1230066 | 9276 | 0.008 | 0.2209 | No | ||
| 106 | CDH16 | 1450129 | 9347 | 0.007 | 0.2171 | No | ||
| 107 | CATSPER2 | 2940403 | 9590 | 0.004 | 0.2040 | No | ||
| 108 | AUH | 5570152 | 9691 | 0.002 | 0.1986 | No | ||
| 109 | RGS7 | 3390044 | 9716 | 0.001 | 0.1973 | No | ||
| 110 | RPH3A | 2190156 | 9738 | 0.001 | 0.1962 | No | ||
| 111 | MTX2 | 6660288 | 9791 | 0.000 | 0.1934 | No | ||
| 112 | KCNK1 | 1050068 | 9819 | -0.000 | 0.1919 | No | ||
| 113 | HMGB3 | 2940168 | 9954 | -0.003 | 0.1847 | No | ||
| 114 | TIMM9 | 2450484 | 10266 | -0.007 | 0.1679 | No | ||
| 115 | ABHD3 | 2370091 | 10320 | -0.008 | 0.1651 | No | ||
| 116 | DIRAS1 | 2640270 | 10497 | -0.011 | 0.1556 | No | ||
| 117 | HCN1 | 5130458 | 10927 | -0.017 | 0.1325 | No | ||
| 118 | CX3CL1 | 3990707 | 11023 | -0.019 | 0.1274 | No | ||
| 119 | SLC30A3 | 1850451 | 11066 | -0.020 | 0.1253 | No | ||
| 120 | KCNMB2 | 3780128 4760136 | 11190 | -0.022 | 0.1188 | No | ||
| 121 | NNAT | 4920053 5130253 | 11606 | -0.030 | 0.0965 | No | ||
| 122 | STK32A | 5890332 | 11640 | -0.031 | 0.0949 | No | ||
| 123 | YWHAH | 1660133 2810053 | 12447 | -0.049 | 0.0516 | No | ||
| 124 | HOXC13 | 4070722 | 12451 | -0.049 | 0.0517 | No | ||
| 125 | TCF2 | 870338 5050632 | 12551 | -0.051 | 0.0467 | No | ||
| 126 | XYLT1 | 6550403 | 12583 | -0.052 | 0.0454 | No | ||
| 127 | CDKL5 | 2450070 | 13048 | -0.067 | 0.0207 | No | ||
| 128 | HCN4 | 1050026 | 13187 | -0.073 | 0.0138 | No | ||
| 129 | RHCG | 6550309 | 13452 | -0.084 | 0.0000 | No | ||
| 130 | ELAVL3 | 2850014 | 13765 | -0.102 | -0.0162 | No | ||
| 131 | MASP1 | 1780619 2900066 | 13773 | -0.103 | -0.0158 | No | ||
| 132 | PNOC | 1990114 | 13837 | -0.108 | -0.0185 | No | ||
| 133 | PHACTR3 | 3850435 5900445 | 13838 | -0.108 | -0.0178 | No | ||
| 134 | VSNL1 | 3120017 | 14068 | -0.125 | -0.0293 | No | ||
| 135 | KIF5A | 510435 | 14128 | -0.131 | -0.0316 | No | ||
| 136 | ASTN2 | 2570136 6940040 | 14134 | -0.131 | -0.0310 | No | ||
| 137 | DEXI | 6220358 6550685 | 14222 | -0.139 | -0.0347 | No | ||
| 138 | TUBB4 | 5310681 | 14382 | -0.154 | -0.0423 | No | ||
| 139 | ATP6V0C | 1780609 | 14595 | -0.182 | -0.0525 | No | ||
| 140 | GBAS | 6760278 | 14707 | -0.198 | -0.0571 | No | ||
| 141 | INSR | 1190504 | 14759 | -0.209 | -0.0584 | No | ||
| 142 | MAEA | 1170392 | 14764 | -0.210 | -0.0572 | No | ||
| 143 | MAL | 4280487 4590239 | 14839 | -0.226 | -0.0596 | No | ||
| 144 | SV2A | 1450039 5570685 | 14955 | -0.252 | -0.0641 | No | ||
| 145 | MEF2D | 5690576 | 15003 | -0.266 | -0.0648 | No | ||
| 146 | SLC9A7 | 5360301 | 15286 | -0.351 | -0.0777 | No | ||
| 147 | RANBP1 | 430215 1090180 | 15436 | -0.407 | -0.0829 | No | ||
| 148 | MTCH2 | 3780603 6620215 | 15458 | -0.418 | -0.0812 | No | ||
| 149 | ATP5B | 3870138 | 15483 | -0.427 | -0.0795 | No | ||
| 150 | MNT | 4920575 | 15674 | -0.510 | -0.0863 | No | ||
| 151 | SEMA3F | 5420601 6770731 | 15698 | -0.520 | -0.0839 | No | ||
| 152 | ZNF289 | 3610390 3870711 3940138 5340373 | 15718 | -0.530 | -0.0813 | No | ||
| 153 | COX4I1 | 5340427 | 15825 | -0.579 | -0.0830 | No | ||
| 154 | GABPA | 2360017 4780301 4850279 | 15846 | -0.588 | -0.0800 | No | ||
| 155 | PANK1 | 670091 6220605 | 16250 | -0.843 | -0.0960 | No | ||
| 156 | PPP3CC | 2450139 | 16390 | -0.935 | -0.0970 | No | ||
| 157 | MSI2 | 1190066 5270039 | 16423 | -0.954 | -0.0921 | No | ||
| 158 | VASP | 7050500 | 16479 | -0.992 | -0.0882 | No | ||
| 159 | UBE1 | 4150139 7050463 | 16518 | -1.011 | -0.0833 | No | ||
| 160 | ESRRA | 6550242 | 16823 | -1.271 | -0.0909 | No | ||
| 161 | VAMP2 | 2100100 | 17100 | -1.571 | -0.0950 | No | ||
| 162 | CHD2 | 4070039 | 17182 | -1.689 | -0.0876 | No | ||
| 163 | IMMT | 870450 4280438 | 17227 | -1.732 | -0.0780 | No | ||
| 164 | MGST3 | 3450338 5290736 | 17445 | -2.055 | -0.0755 | No | ||
| 165 | CRSP7 | 4230408 | 17535 | -2.184 | -0.0652 | No | ||
| 166 | FBXO18 | 1240731 6370541 | 17764 | -2.618 | -0.0593 | No | ||
| 167 | ARF3 | 3710446 | 17803 | -2.693 | -0.0427 | No | ||
| 168 | ATP1B1 | 3130594 | 18145 | -3.686 | -0.0356 | No | ||
| 169 | RANBP10 | 2340184 | 18203 | -3.900 | -0.0116 | No | ||
| 170 | GABARAPL1 | 2810458 | 18376 | -4.892 | 0.0130 | No |