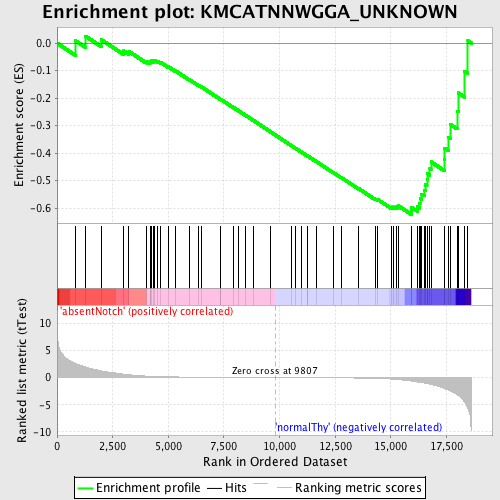
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | KMCATNNWGGA_UNKNOWN |
| Enrichment Score (ES) | -0.6229764 |
| Normalized Enrichment Score (NES) | -1.5010338 |
| Nominal p-value | 0.0060728746 |
| FDR q-value | 0.1475709 |
| FWER p-Value | 0.807 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CABP1 | 6350750 | 828 | 2.564 | 0.0087 | No | ||
| 2 | ESM1 | 1410594 | 1276 | 1.966 | 0.0255 | No | ||
| 3 | BCL2L13 | 3170593 | 1993 | 1.224 | 0.0124 | No | ||
| 4 | ABL1 | 1050593 2030050 4010114 | 2989 | 0.624 | -0.0283 | No | ||
| 5 | NR2F1 | 4120528 360402 | 3230 | 0.514 | -0.0305 | No | ||
| 6 | SOX12 | 1780037 | 4007 | 0.275 | -0.0666 | No | ||
| 7 | CNOT3 | 130519 4480292 1990609 | 4177 | 0.240 | -0.0707 | No | ||
| 8 | STAG1 | 1190300 1400722 4590100 | 4206 | 0.235 | -0.0673 | No | ||
| 9 | UFD1L | 870020 | 4242 | 0.231 | -0.0644 | No | ||
| 10 | CAB39L | 5670040 | 4349 | 0.214 | -0.0657 | No | ||
| 11 | NR2F2 | 3170609 3310577 | 4356 | 0.213 | -0.0616 | No | ||
| 12 | PDE4D | 2470528 6660014 | 4523 | 0.188 | -0.0666 | No | ||
| 13 | PNPT1 | 5270433 | 4659 | 0.171 | -0.0703 | No | ||
| 14 | SYNCRIP | 1690195 3140113 4670279 | 5008 | 0.136 | -0.0863 | No | ||
| 15 | ENAH | 1690292 5700300 | 5326 | 0.112 | -0.1010 | No | ||
| 16 | SOCS5 | 3830398 7100093 | 5957 | 0.080 | -0.1333 | No | ||
| 17 | ADAMTSL1 | 2970092 | 6373 | 0.066 | -0.1543 | No | ||
| 18 | KCNJ9 | 3440292 | 6474 | 0.063 | -0.1584 | No | ||
| 19 | CSNK1A1 | 2340427 | 7327 | 0.041 | -0.2034 | No | ||
| 20 | SFRS1 | 2360440 | 7351 | 0.041 | -0.2038 | No | ||
| 21 | SP3 | 3840338 | 7948 | 0.029 | -0.2353 | No | ||
| 22 | IL2 | 1770725 | 8133 | 0.026 | -0.2447 | No | ||
| 23 | SNX5 | 1570021 6860609 | 8484 | 0.020 | -0.2632 | No | ||
| 24 | RBM16 | 6290286 6400181 | 8845 | 0.014 | -0.2823 | No | ||
| 25 | ZBTB11 | 6520092 | 9578 | 0.004 | -0.3216 | No | ||
| 26 | PAX6 | 1190025 | 10551 | -0.011 | -0.3738 | No | ||
| 27 | LMO1 | 3170528 | 10732 | -0.014 | -0.3832 | No | ||
| 28 | TSC1 | 1850672 | 10977 | -0.018 | -0.3959 | No | ||
| 29 | PPP4R2 | 5890035 | 11234 | -0.023 | -0.4093 | No | ||
| 30 | DMD | 1740041 3990332 | 11254 | -0.023 | -0.4098 | No | ||
| 31 | SDCCAG10 | 2630286 | 11662 | -0.031 | -0.4311 | No | ||
| 32 | CELSR3 | 5270731 7100082 | 12409 | -0.048 | -0.4703 | No | ||
| 33 | OSBPL6 | 3800072 | 12778 | -0.058 | -0.4889 | No | ||
| 34 | GAS7 | 2120358 6450494 | 13563 | -0.091 | -0.5293 | No | ||
| 35 | RECK | 840079 | 14321 | -0.149 | -0.5670 | No | ||
| 36 | DUSP7 | 2940309 | 14406 | -0.157 | -0.5682 | No | ||
| 37 | EPHA2 | 5890056 | 15022 | -0.269 | -0.5958 | No | ||
| 38 | CRAT | 540020 2060364 3520148 | 15138 | -0.300 | -0.5957 | No | ||
| 39 | CDC45L | 70537 3130114 | 15261 | -0.345 | -0.5951 | No | ||
| 40 | SPRED2 | 510138 540195 | 15343 | -0.370 | -0.5918 | No | ||
| 41 | STRN4 | 3440711 | 15923 | -0.629 | -0.6099 | Yes | ||
| 42 | CRMP1 | 4810014 | 15934 | -0.632 | -0.5973 | Yes | ||
| 43 | AP1G1 | 2360671 | 16207 | -0.803 | -0.5952 | Yes | ||
| 44 | BRUNOL4 | 4490452 5080451 | 16298 | -0.875 | -0.5819 | Yes | ||
| 45 | ING3 | 110438 2060390 | 16314 | -0.886 | -0.5643 | Yes | ||
| 46 | ANAPC7 | 5260274 | 16389 | -0.934 | -0.5488 | Yes | ||
| 47 | CGGBP1 | 4810053 | 16516 | -1.010 | -0.5346 | Yes | ||
| 48 | GEMIN4 | 130278 2230075 | 16557 | -1.033 | -0.5153 | Yes | ||
| 49 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 16627 | -1.087 | -0.4964 | Yes | ||
| 50 | MAP4K3 | 5390551 | 16648 | -1.104 | -0.4745 | Yes | ||
| 51 | AEBP2 | 2970603 | 16756 | -1.208 | -0.4551 | Yes | ||
| 52 | MAP2K5 | 2030440 | 16809 | -1.262 | -0.4317 | Yes | ||
| 53 | HIC2 | 4010433 | 17404 | -1.988 | -0.4223 | Yes | ||
| 54 | WASL | 3290170 | 17427 | -2.029 | -0.3813 | Yes | ||
| 55 | FKRP | 130546 | 17583 | -2.285 | -0.3421 | Yes | ||
| 56 | RSHL2 | 6110014 | 17690 | -2.473 | -0.2964 | Yes | ||
| 57 | DUSP6 | 5910286 7100070 | 17982 | -3.129 | -0.2470 | Yes | ||
| 58 | HOXA7 | 5910152 | 18035 | -3.305 | -0.1811 | Yes | ||
| 59 | MRPL24 | 1170014 | 18316 | -4.526 | -0.1020 | Yes | ||
| 60 | ARNTL | 3170463 | 18464 | -5.680 | 0.0082 | Yes |

