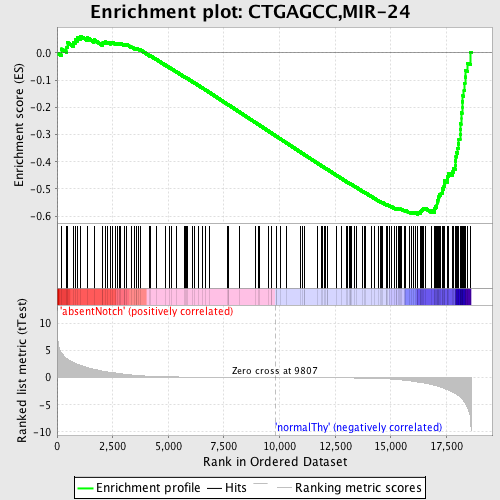
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CTGAGCC,MIR-24 |
| Enrichment Score (ES) | -0.5938943 |
| Normalized Enrichment Score (NES) | -1.6215849 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09997225 |
| FWER p-Value | 0.229 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ABCB9 | 110603 4560592 6110338 | 195 | 4.554 | 0.0147 | No | ||
| 2 | MAFG | 4120440 | 435 | 3.487 | 0.0212 | No | ||
| 3 | CCT3 | 7100403 | 461 | 3.376 | 0.0386 | No | ||
| 4 | PPM1G | 610725 | 740 | 2.735 | 0.0387 | No | ||
| 5 | AARS | 1570279 2370136 | 811 | 2.592 | 0.0493 | No | ||
| 6 | GPX3 | 1340450 | 916 | 2.431 | 0.0572 | No | ||
| 7 | PER1 | 3290315 | 1057 | 2.234 | 0.0620 | No | ||
| 8 | B3GNT6 | 1340736 | 1357 | 1.869 | 0.0562 | No | ||
| 9 | RNF11 | 3990068 5080458 | 1659 | 1.534 | 0.0484 | No | ||
| 10 | NEK9 | 3440402 | 2058 | 1.175 | 0.0334 | No | ||
| 11 | RAP1B | 3710138 | 2060 | 1.174 | 0.0399 | No | ||
| 12 | CMTM4 | 360600 3520332 | 2152 | 1.095 | 0.0410 | No | ||
| 13 | FBXL19 | 4570039 | 2274 | 1.017 | 0.0401 | No | ||
| 14 | KPNA3 | 7040088 | 2411 | 0.963 | 0.0381 | No | ||
| 15 | CDKN1B | 3800025 6450044 | 2494 | 0.906 | 0.0387 | No | ||
| 16 | XPO4 | 3840551 | 2631 | 0.824 | 0.0359 | No | ||
| 17 | SLC19A2 | 6200750 | 2712 | 0.770 | 0.0358 | No | ||
| 18 | PIGS | 3610142 | 2800 | 0.719 | 0.0351 | No | ||
| 19 | STC2 | 4920601 | 2854 | 0.691 | 0.0361 | No | ||
| 20 | PARP6 | 3290332 4760167 6900181 | 3025 | 0.608 | 0.0303 | No | ||
| 21 | RASGRP4 | 540102 | 3039 | 0.603 | 0.0329 | No | ||
| 22 | CDV3 | 1940577 | 3097 | 0.572 | 0.0330 | No | ||
| 23 | DLGAP4 | 4570672 5690161 | 3346 | 0.469 | 0.0222 | No | ||
| 24 | MMP14 | 670079 | 3491 | 0.419 | 0.0167 | No | ||
| 25 | TNFRSF19 | 3870731 6940730 | 3549 | 0.399 | 0.0158 | No | ||
| 26 | ACVR1B | 3610446 5570195 | 3585 | 0.386 | 0.0161 | No | ||
| 27 | RAP2C | 1690132 | 3675 | 0.360 | 0.0133 | No | ||
| 28 | PDGFRA | 2940332 | 3734 | 0.341 | 0.0120 | No | ||
| 29 | EDA | 6420441 | 4172 | 0.242 | -0.0103 | No | ||
| 30 | NDST1 | 1500427 5340121 | 4218 | 0.233 | -0.0115 | No | ||
| 31 | TRIM33 | 580619 2230280 3990433 6200747 | 4472 | 0.196 | -0.0241 | No | ||
| 32 | FGFR3 | 5390632 6020021 | 4852 | 0.149 | -0.0438 | No | ||
| 33 | FST | 1110600 | 5070 | 0.131 | -0.0548 | No | ||
| 34 | MXI1 | 5050064 5130484 | 5122 | 0.127 | -0.0569 | No | ||
| 35 | AMMECR1 | 3800435 | 5346 | 0.110 | -0.0683 | No | ||
| 36 | NEK4 | 430731 2060010 7000333 | 5729 | 0.090 | -0.0885 | No | ||
| 37 | RNF150 | 7000500 | 5771 | 0.088 | -0.0903 | No | ||
| 38 | SEMA6B | 6770725 7100091 | 5829 | 0.085 | -0.0929 | No | ||
| 39 | SMCR8 | 730653 | 5851 | 0.084 | -0.0935 | No | ||
| 40 | PRSS8 | 3170577 | 6097 | 0.075 | -0.1064 | No | ||
| 41 | PTGER4 | 6400670 | 6186 | 0.072 | -0.1108 | No | ||
| 42 | KLHL1 | 6350021 | 6337 | 0.067 | -0.1185 | No | ||
| 43 | POGZ | 870348 7040286 | 6538 | 0.060 | -0.1290 | No | ||
| 44 | NELL2 | 2340725 4280301 5420008 | 6676 | 0.057 | -0.1361 | No | ||
| 45 | BHLHB5 | 6510520 | 6867 | 0.051 | -0.1461 | No | ||
| 46 | JUB | 5860711 5900500 | 7666 | 0.034 | -0.1892 | No | ||
| 47 | KCNK2 | 1230592 7000497 | 7690 | 0.034 | -0.1902 | No | ||
| 48 | BSN | 5270347 | 7720 | 0.033 | -0.1916 | No | ||
| 49 | RAP1A | 1090025 | 8179 | 0.025 | -0.2163 | No | ||
| 50 | EDG1 | 4200619 | 8896 | 0.014 | -0.2550 | No | ||
| 51 | TRPM6 | 2570072 | 9054 | 0.011 | -0.2634 | No | ||
| 52 | CALCR | 1690494 | 9113 | 0.010 | -0.2665 | No | ||
| 53 | EXOC5 | 7000711 | 9493 | 0.005 | -0.2870 | No | ||
| 54 | MMP16 | 2680139 | 9619 | 0.003 | -0.2938 | No | ||
| 55 | AMOTL2 | 4730082 | 9655 | 0.002 | -0.2957 | No | ||
| 56 | GPC4 | 5910408 | 9860 | -0.001 | -0.3067 | No | ||
| 57 | PURA | 3870156 | 9881 | -0.001 | -0.3078 | No | ||
| 58 | RARG | 6760136 | 10018 | -0.004 | -0.3151 | No | ||
| 59 | MAPK7 | 5670079 6100128 | 10299 | -0.008 | -0.3303 | No | ||
| 60 | ARID5B | 110403 | 10925 | -0.017 | -0.3640 | No | ||
| 61 | YBX2 | 3520441 | 11014 | -0.019 | -0.3687 | No | ||
| 62 | NEUROD1 | 3060619 | 11111 | -0.021 | -0.3738 | No | ||
| 63 | EPHA8 | 6520242 | 11141 | -0.021 | -0.3752 | No | ||
| 64 | LYPLA2 | 1500112 | 11697 | -0.032 | -0.4051 | No | ||
| 65 | APBA1 | 3140487 | 11905 | -0.036 | -0.4161 | No | ||
| 66 | APOA5 | 6130471 | 11918 | -0.036 | -0.4166 | No | ||
| 67 | PCDH10 | 940014 1190592 3190204 | 12038 | -0.039 | -0.4228 | No | ||
| 68 | DOCK3 | 3290273 3360497 2850068 | 12077 | -0.040 | -0.4246 | No | ||
| 69 | SFXN5 | 2630152 | 12151 | -0.042 | -0.4284 | No | ||
| 70 | SCML2 | 3290603 | 12161 | -0.042 | -0.4286 | No | ||
| 71 | TCF2 | 870338 5050632 | 12551 | -0.051 | -0.4494 | No | ||
| 72 | DLC1 | 1090632 6450594 | 12770 | -0.058 | -0.4609 | No | ||
| 73 | TMEM9 | 130039 | 13023 | -0.066 | -0.4742 | No | ||
| 74 | MEN1 | 4850300 5900671 | 13065 | -0.068 | -0.4760 | No | ||
| 75 | CLCN6 | 3520400 | 13151 | -0.071 | -0.4802 | No | ||
| 76 | LRFN2 | 6450411 | 13179 | -0.073 | -0.4813 | No | ||
| 77 | IGFBP5 | 2360592 | 13190 | -0.073 | -0.4814 | No | ||
| 78 | KIF21B | 2630026 6200020 6980040 | 13249 | -0.075 | -0.4841 | No | ||
| 79 | PTPN9 | 3290408 | 13374 | -0.081 | -0.4904 | No | ||
| 80 | BCL2L2 | 2760692 6770739 | 13474 | -0.085 | -0.4953 | No | ||
| 81 | SP6 | 60484 510452 2690333 | 13741 | -0.101 | -0.5092 | No | ||
| 82 | COL11A2 | 5130341 | 13822 | -0.107 | -0.5129 | No | ||
| 83 | LENG4 | 2060400 5700487 | 13878 | -0.111 | -0.5153 | No | ||
| 84 | DCBLD2 | 5890333 | 14122 | -0.130 | -0.5277 | No | ||
| 85 | CDX2 | 3850349 | 14264 | -0.144 | -0.5345 | No | ||
| 86 | CNTFR | 4480092 6200064 | 14463 | -0.163 | -0.5444 | No | ||
| 87 | PTPRD | 520008 3120097 | 14550 | -0.173 | -0.5481 | No | ||
| 88 | DUSP16 | 2370082 2480450 | 14591 | -0.180 | -0.5492 | No | ||
| 89 | INSIG1 | 1500332 3450458 4540301 | 14641 | -0.189 | -0.5508 | No | ||
| 90 | RNF2 | 2340497 | 14816 | -0.220 | -0.5590 | No | ||
| 91 | FURIN | 4120168 | 14823 | -0.222 | -0.5581 | No | ||
| 92 | GRIA3 | 2900164 6130278 6860154 | 14855 | -0.228 | -0.5585 | No | ||
| 93 | ING5 | 3830440 5910239 6200471 6650576 | 14943 | -0.249 | -0.5619 | No | ||
| 94 | SP1 | 6590017 | 15040 | -0.275 | -0.5655 | No | ||
| 95 | CACNB1 | 2940427 3710487 | 15174 | -0.314 | -0.5710 | No | ||
| 96 | BCL2L11 | 780044 4200601 | 15249 | -0.339 | -0.5731 | No | ||
| 97 | ADCY9 | 5690168 | 15263 | -0.345 | -0.5719 | No | ||
| 98 | MATR3 | 1940170 5340278 | 15354 | -0.373 | -0.5747 | No | ||
| 99 | PLOD2 | 3360427 | 15397 | -0.390 | -0.5748 | No | ||
| 100 | MAP3K7IP2 | 2340242 | 15413 | -0.395 | -0.5734 | No | ||
| 101 | INPP5B | 6370088 | 15486 | -0.428 | -0.5749 | No | ||
| 102 | SYP | 2690168 | 15602 | -0.474 | -0.5785 | No | ||
| 103 | MNT | 4920575 | 15674 | -0.510 | -0.5796 | No | ||
| 104 | MARK4 | 730519 | 15838 | -0.585 | -0.5851 | No | ||
| 105 | SLC12A6 | 610278 1980035 5340671 | 15937 | -0.634 | -0.5869 | No | ||
| 106 | PHC1 | 520195 | 16006 | -0.677 | -0.5868 | No | ||
| 107 | ZXDA | 1580332 | 16087 | -0.720 | -0.5872 | No | ||
| 108 | WDTC1 | 1690541 | 16212 | -0.810 | -0.5894 | Yes | ||
| 109 | TAF5 | 3450288 5890193 6860435 | 16218 | -0.813 | -0.5851 | Yes | ||
| 110 | HAP1 | 1240010 4230079 6980195 | 16324 | -0.895 | -0.5859 | Yes | ||
| 111 | DNAJB12 | 2030148 | 16336 | -0.901 | -0.5815 | Yes | ||
| 112 | TOP1 | 770471 4060632 6650324 | 16388 | -0.933 | -0.5790 | Yes | ||
| 113 | RAB5B | 130397 1980273 6900731 | 16415 | -0.947 | -0.5752 | Yes | ||
| 114 | PER2 | 3120204 3390593 | 16454 | -0.980 | -0.5718 | Yes | ||
| 115 | MAGI1 | 6510500 1990239 4670471 | 16539 | -1.026 | -0.5706 | Yes | ||
| 116 | DHX30 | 2480494 | 16810 | -1.263 | -0.5783 | Yes | ||
| 117 | OGT | 2360131 4610333 | 16965 | -1.415 | -0.5787 | Yes | ||
| 118 | SUV420H2 | 4050368 | 16974 | -1.422 | -0.5713 | Yes | ||
| 119 | PLK3 | 2640592 | 17013 | -1.465 | -0.5652 | Yes | ||
| 120 | PAK4 | 1690152 | 17070 | -1.536 | -0.5597 | Yes | ||
| 121 | RNF138 | 110142 3870129 6020441 6400193 | 17075 | -1.543 | -0.5513 | Yes | ||
| 122 | ATP6V0A2 | 4050129 | 17084 | -1.552 | -0.5431 | Yes | ||
| 123 | SBK1 | 940452 | 17121 | -1.602 | -0.5362 | Yes | ||
| 124 | SSH2 | 360056 | 17159 | -1.657 | -0.5290 | Yes | ||
| 125 | RASA1 | 1240315 | 17172 | -1.670 | -0.5204 | Yes | ||
| 126 | RASSF2 | 2480079 5720546 | 17254 | -1.776 | -0.5149 | Yes | ||
| 127 | CSK | 6350593 | 17311 | -1.836 | -0.5077 | Yes | ||
| 128 | PIM2 | 840136 5720692 7100577 | 17317 | -1.840 | -0.4978 | Yes | ||
| 129 | RHBDL1 | 1500736 | 17386 | -1.956 | -0.4906 | Yes | ||
| 130 | HIC2 | 4010433 | 17404 | -1.988 | -0.4805 | Yes | ||
| 131 | RAB5C | 1990397 | 17418 | -2.009 | -0.4700 | Yes | ||
| 132 | WDR23 | 2030372 | 17534 | -2.184 | -0.4641 | Yes | ||
| 133 | PIM1 | 630047 | 17549 | -2.218 | -0.4525 | Yes | ||
| 134 | PRKRIP1 | 4280746 6100390 | 17602 | -2.319 | -0.4425 | Yes | ||
| 135 | H2AFX | 3520082 | 17782 | -2.659 | -0.4374 | Yes | ||
| 136 | ARHGEF5 | 3170605 | 17804 | -2.693 | -0.4236 | Yes | ||
| 137 | CLK2 | 510079 2630736 | 17896 | -2.903 | -0.4124 | Yes | ||
| 138 | PTPRF | 1770528 3190044 | 17906 | -2.921 | -0.3966 | Yes | ||
| 139 | MIDN | 2260132 5220377 | 17925 | -2.959 | -0.3812 | Yes | ||
| 140 | PLCL2 | 6100575 | 17953 | -3.038 | -0.3657 | Yes | ||
| 141 | HIP1R | 3120368 | 18006 | -3.210 | -0.3507 | Yes | ||
| 142 | MOCS1 | 2120022 | 18038 | -3.309 | -0.3340 | Yes | ||
| 143 | ACBD4 | 6980079 | 18058 | -3.369 | -0.3163 | Yes | ||
| 144 | PIP5K1B | 3450113 6380278 | 18112 | -3.567 | -0.2994 | Yes | ||
| 145 | NEDD9 | 1740373 4230053 | 18125 | -3.600 | -0.2800 | Yes | ||
| 146 | VAV1 | 6020487 | 18137 | -3.653 | -0.2603 | Yes | ||
| 147 | SESN1 | 2120021 | 18157 | -3.737 | -0.2406 | Yes | ||
| 148 | ADAM19 | 5340075 | 18159 | -3.747 | -0.2198 | Yes | ||
| 149 | RANBP10 | 2340184 | 18203 | -3.900 | -0.2005 | Yes | ||
| 150 | RGL2 | 1740017 | 18207 | -3.929 | -0.1788 | Yes | ||
| 151 | ALS2CR2 | 6450128 7100092 | 18238 | -4.069 | -0.1578 | Yes | ||
| 152 | GBA2 | 2900372 | 18288 | -4.338 | -0.1364 | Yes | ||
| 153 | TOB2 | 1240465 | 18313 | -4.507 | -0.1126 | Yes | ||
| 154 | PSAP | 2810338 | 18336 | -4.615 | -0.0882 | Yes | ||
| 155 | RAB6IP1 | 4150044 | 18362 | -4.773 | -0.0630 | Yes | ||
| 156 | SEMA4A | 5890022 | 18455 | -5.594 | -0.0369 | Yes | ||
| 157 | TRPC4AP | 840520 1660402 | 18588 | -8.204 | 0.0015 | Yes |

