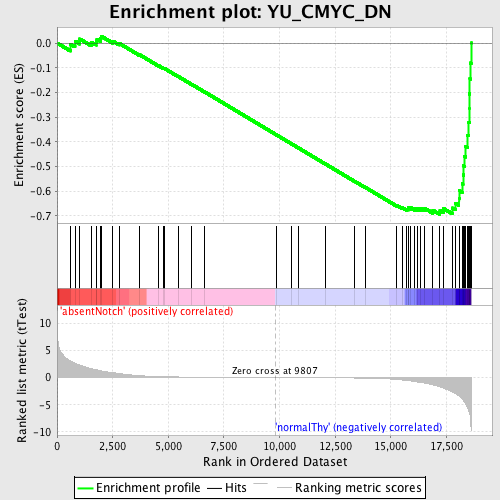
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | YU_CMYC_DN |
| Enrichment Score (ES) | -0.6940344 |
| Normalized Enrichment Score (NES) | -1.6168609 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.258446 |
| FWER p-Value | 0.883 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UNC93B1 | 430152 | 600 | 3.038 | -0.0050 | No | ||
| 2 | CTSH | 60524 | 805 | 2.610 | 0.0074 | No | ||
| 3 | IGH-1A | 360113 4670341 | 1019 | 2.276 | 0.0164 | No | ||
| 4 | PTPRC | 130402 5290148 | 1532 | 1.647 | 0.0036 | No | ||
| 5 | H2-AA | 5290673 | 1750 | 1.445 | 0.0049 | No | ||
| 6 | GNS | 3120458 | 1789 | 1.409 | 0.0155 | No | ||
| 7 | H2-AB1 | 4230427 | 1950 | 1.265 | 0.0182 | No | ||
| 8 | H2-DMB1 | 4760026 | 1976 | 1.237 | 0.0280 | No | ||
| 9 | GDI1 | 4590400 5290358 6020687 6180446 | 2502 | 0.902 | 0.0078 | No | ||
| 10 | RBMS1 | 6400014 | 2796 | 0.721 | -0.0015 | No | ||
| 11 | H2-EB1 | 2350079 | 3697 | 0.352 | -0.0468 | No | ||
| 12 | OLFM1 | 3360161 5570066 | 4563 | 0.183 | -0.0918 | No | ||
| 13 | MACF1 | 1340132 2370176 2900750 6590364 | 4787 | 0.155 | -0.1024 | No | ||
| 14 | SRPK3 | 2480278 | 4804 | 0.154 | -0.1019 | No | ||
| 15 | GAD1 | 2360035 3140167 | 5444 | 0.104 | -0.1354 | No | ||
| 16 | PYGM | 3940300 | 6035 | 0.078 | -0.1665 | No | ||
| 17 | LOC207685 | 6520017 | 6605 | 0.059 | -0.1966 | No | ||
| 18 | ABCA1 | 6290156 | 9882 | -0.001 | -0.3730 | No | ||
| 19 | 5730420B22RIK | 130139 | 10530 | -0.011 | -0.4078 | No | ||
| 20 | SSPN | 4670091 | 10865 | -0.017 | -0.4256 | No | ||
| 21 | MYO7A | 2810102 | 12051 | -0.039 | -0.4891 | No | ||
| 22 | GGA2 | 520451 1240082 2650332 | 13386 | -0.081 | -0.5602 | No | ||
| 23 | MS4A1 | 4210598 | 13865 | -0.110 | -0.5850 | No | ||
| 24 | 1100001G20RIK | 3610368 | 15259 | -0.342 | -0.6570 | No | ||
| 25 | IFI203 | 6420102 3610369 | 15502 | -0.433 | -0.6661 | No | ||
| 26 | ACP5 | 2230717 | 15717 | -0.529 | -0.6729 | No | ||
| 27 | MGST1 | 6020605 | 15774 | -0.556 | -0.6709 | No | ||
| 28 | H2-DMA | 6290463 | 15791 | -0.564 | -0.6667 | No | ||
| 29 | GBP4 | 6520471 1980670 | 15897 | -0.617 | -0.6668 | No | ||
| 30 | LAPTM5 | 5550372 | 16079 | -0.715 | -0.6701 | No | ||
| 31 | CMAH | 1660497 3390398 6550040 | 16216 | -0.812 | -0.6702 | No | ||
| 32 | PRKCB1 | 870019 3130092 | 16350 | -0.909 | -0.6692 | No | ||
| 33 | BLK | 1940128 5390053 | 16526 | -1.017 | -0.6695 | No | ||
| 34 | H2-OB | 6980059 | 16890 | -1.343 | -0.6770 | No | ||
| 35 | SIPA1 | 5220687 | 17208 | -1.715 | -0.6786 | Yes | ||
| 36 | DGKA | 5720152 5890328 | 17359 | -1.901 | -0.6696 | Yes | ||
| 37 | RAG1 | 2850279 | 17758 | -2.607 | -0.6677 | Yes | ||
| 38 | CD2 | 430672 | 17910 | -2.930 | -0.6495 | Yes | ||
| 39 | IL4I1 | 3190161 | 18064 | -3.382 | -0.6274 | Yes | ||
| 40 | EVL | 1740113 | 18072 | -3.430 | -0.5970 | Yes | ||
| 41 | RGL2 | 1740017 | 18207 | -3.929 | -0.5689 | Yes | ||
| 42 | CD37 | 770273 | 18246 | -4.132 | -0.5338 | Yes | ||
| 43 | KLF2 | 6860270 | 18275 | -4.251 | -0.4972 | Yes | ||
| 44 | KLF3 | 5130438 | 18314 | -4.509 | -0.4587 | Yes | ||
| 45 | CD1D1 | 5130184 | 18369 | -4.863 | -0.4180 | Yes | ||
| 46 | ARHGEF1 | 610347 4850603 6420672 | 18456 | -5.598 | -0.3724 | Yes | ||
| 47 | LTB | 3990672 | 18503 | -6.099 | -0.3201 | Yes | ||
| 48 | IL10RA | 1770164 | 18528 | -6.413 | -0.2638 | Yes | ||
| 49 | MAP4K2 | 4200037 | 18533 | -6.506 | -0.2056 | Yes | ||
| 50 | SLFN2 | 3120114 | 18555 | -6.844 | -0.1453 | Yes | ||
| 51 | ARID3B | 6650008 | 18571 | -7.279 | -0.0807 | Yes | ||
| 52 | BTG1 | 4200735 6040131 6200133 | 18611 | -9.256 | 0.0003 | Yes |

