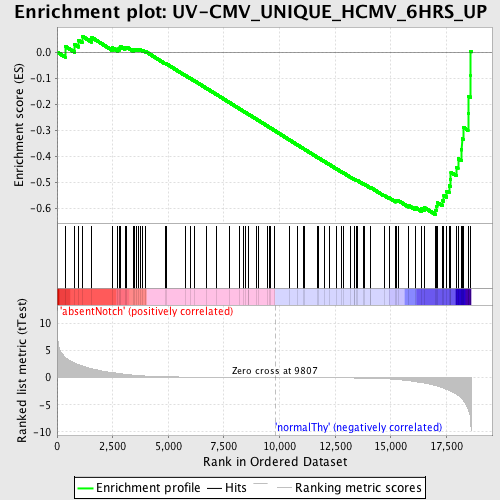
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | UV-CMV_UNIQUE_HCMV_6HRS_UP |
| Enrichment Score (ES) | -0.6220956 |
| Normalized Enrichment Score (NES) | -1.5794816 |
| Nominal p-value | 0.0018975332 |
| FDR q-value | 0.26476142 |
| FWER p-Value | 0.986 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MX2 | 2030373 | 363 | 3.705 | 0.0219 | No | ||
| 2 | PIK3R3 | 1770333 | 789 | 2.640 | 0.0285 | No | ||
| 3 | CCL8 | 3870010 | 966 | 2.341 | 0.0452 | No | ||
| 4 | PPP1R16B | 3520300 | 1125 | 2.158 | 0.0608 | No | ||
| 5 | CSTF2 | 6040463 | 1550 | 1.631 | 0.0562 | No | ||
| 6 | RQCD1 | 1850139 | 2479 | 0.916 | 0.0164 | No | ||
| 7 | STX11 | 2470687 3710528 6590184 | 2727 | 0.763 | 0.0116 | No | ||
| 8 | CX3CR1 | 1690577 | 2806 | 0.715 | 0.0154 | No | ||
| 9 | PFKFB2 | 3940538 7100059 | 2846 | 0.699 | 0.0211 | No | ||
| 10 | TNFSF10 | 3170300 | 3052 | 0.596 | 0.0167 | No | ||
| 11 | SAMHD1 | 2350338 5390451 | 3128 | 0.555 | 0.0189 | No | ||
| 12 | CH25H | 4210167 | 3419 | 0.442 | 0.0082 | No | ||
| 13 | RBBP6 | 2320129 | 3493 | 0.418 | 0.0090 | No | ||
| 14 | ING1 | 5690010 6520056 | 3562 | 0.394 | 0.0097 | No | ||
| 15 | EGR3 | 6940128 | 3667 | 0.363 | 0.0081 | No | ||
| 16 | ELN | 5080347 | 3727 | 0.343 | 0.0088 | No | ||
| 17 | TAF13 | 4050692 | 3830 | 0.315 | 0.0068 | No | ||
| 18 | BRCA2 | 4280372 | 3965 | 0.282 | 0.0027 | No | ||
| 19 | FGFR3 | 5390632 6020021 | 4852 | 0.149 | -0.0434 | No | ||
| 20 | KIF5C | 6370717 | 4923 | 0.143 | -0.0456 | No | ||
| 21 | EPHA5 | 2340671 | 5773 | 0.088 | -0.0904 | No | ||
| 22 | HBEGF | 2030156 | 5989 | 0.079 | -0.1011 | No | ||
| 23 | IER2 | 2030008 | 6171 | 0.072 | -0.1101 | No | ||
| 24 | PDE1A | 1050458 2570563 | 6728 | 0.055 | -0.1394 | No | ||
| 25 | MEF2C | 670025 780338 | 7167 | 0.044 | -0.1626 | No | ||
| 26 | PCDH9 | 2100136 | 7758 | 0.033 | -0.1941 | No | ||
| 27 | DNAJB9 | 460114 | 8175 | 0.025 | -0.2162 | No | ||
| 28 | GALNT3 | 1850687 | 8364 | 0.022 | -0.2261 | No | ||
| 29 | PLA2G4A | 6380364 | 8459 | 0.021 | -0.2310 | No | ||
| 30 | GEM | 5290082 | 8602 | 0.019 | -0.2384 | No | ||
| 31 | CRY1 | 6770039 | 8613 | 0.019 | -0.2387 | No | ||
| 32 | MSX1 | 2650309 | 8971 | 0.012 | -0.2579 | No | ||
| 33 | TREX1 | 3450040 7100692 | 9057 | 0.011 | -0.2623 | No | ||
| 34 | TFEC | 1170722 | 9434 | 0.006 | -0.2826 | No | ||
| 35 | TLR3 | 6760451 | 9563 | 0.004 | -0.2894 | No | ||
| 36 | BAMBI | 6650463 | 9589 | 0.004 | -0.2907 | No | ||
| 37 | IL6 | 380133 | 9755 | 0.001 | -0.2996 | No | ||
| 38 | TROVE2 | 4850458 | 10423 | -0.010 | -0.3355 | No | ||
| 39 | CRYGD | 2680095 | 10812 | -0.016 | -0.3563 | No | ||
| 40 | COL9A3 | 4050541 | 11086 | -0.020 | -0.3708 | No | ||
| 41 | SOS1 | 7050338 | 11112 | -0.021 | -0.3719 | No | ||
| 42 | AGTPBP1 | 1990735 | 11701 | -0.032 | -0.4033 | No | ||
| 43 | CHEK2 | 610139 1050022 | 11739 | -0.033 | -0.4049 | No | ||
| 44 | CD38 | 6100056 | 12019 | -0.039 | -0.4195 | No | ||
| 45 | TRIM21 | 3360026 | 12247 | -0.044 | -0.4313 | No | ||
| 46 | FGF2 | 3450433 | 12553 | -0.051 | -0.4472 | No | ||
| 47 | DPT | 5670731 | 12780 | -0.059 | -0.4587 | No | ||
| 48 | CPEB3 | 3940164 | 12871 | -0.061 | -0.4629 | No | ||
| 49 | INDO | 2680390 | 13185 | -0.073 | -0.4789 | No | ||
| 50 | TDRD7 | 6550520 | 13357 | -0.080 | -0.4873 | No | ||
| 51 | AREG | 4920025 | 13363 | -0.080 | -0.4866 | No | ||
| 52 | IGFBP2 | 5220563 | 13463 | -0.084 | -0.4910 | No | ||
| 53 | CCL19 | 5080487 | 13512 | -0.087 | -0.4927 | No | ||
| 54 | TAC1 | 7000195 380706 | 13761 | -0.102 | -0.5049 | No | ||
| 55 | NP | 2630577 3120142 5670068 6290039 | 13811 | -0.106 | -0.5064 | No | ||
| 56 | IL1RAP | 50546 450711 1580397 3940301 5890181 6370373 | 14085 | -0.127 | -0.5197 | No | ||
| 57 | EGFR | 4920138 6480521 | 14087 | -0.127 | -0.5183 | No | ||
| 58 | OAS2 | 1230408 | 14696 | -0.197 | -0.5489 | No | ||
| 59 | TNFSF9 | 1190408 | 14927 | -0.244 | -0.5586 | No | ||
| 60 | RGS6 | 6040601 | 15221 | -0.328 | -0.5707 | No | ||
| 61 | ELF1 | 4780450 | 15262 | -0.345 | -0.5690 | No | ||
| 62 | UBE2S | 1850110 | 15322 | -0.364 | -0.5681 | No | ||
| 63 | PTTG1 | 520463 | 15797 | -0.567 | -0.5873 | No | ||
| 64 | GTF2B | 6860685 | 16123 | -0.749 | -0.5965 | No | ||
| 65 | RABGAP1L | 3190014 6860088 | 16376 | -0.922 | -0.5998 | No | ||
| 66 | CREM | 840156 6380438 6660041 6660168 | 16502 | -1.000 | -0.5953 | No | ||
| 67 | IRF7 | 1570605 | 16999 | -1.452 | -0.6058 | Yes | ||
| 68 | IRF4 | 610133 | 17043 | -1.501 | -0.5914 | Yes | ||
| 69 | MYD88 | 2100717 | 17076 | -1.543 | -0.5758 | Yes | ||
| 70 | NFKBIA | 1570152 | 17332 | -1.860 | -0.5688 | Yes | ||
| 71 | CCND2 | 5340167 | 17384 | -1.954 | -0.5496 | Yes | ||
| 72 | RELB | 1400048 | 17503 | -2.129 | -0.5322 | Yes | ||
| 73 | NFKB2 | 2320670 | 17622 | -2.349 | -0.5122 | Yes | ||
| 74 | DUSP2 | 2370184 | 17680 | -2.450 | -0.4879 | Yes | ||
| 75 | ICAM1 | 6980138 | 17701 | -2.507 | -0.4609 | Yes | ||
| 76 | COL15A1 | 6840113 | 17942 | -2.995 | -0.4403 | Yes | ||
| 77 | RRAD | 4280129 | 18022 | -3.260 | -0.4081 | Yes | ||
| 78 | JUNB | 4230048 | 18168 | -3.777 | -0.3737 | Yes | ||
| 79 | ISG20 | 1770750 | 18199 | -3.891 | -0.3317 | Yes | ||
| 80 | SOCS1 | 730139 | 18254 | -4.161 | -0.2881 | Yes | ||
| 81 | EGR2 | 3800403 | 18471 | -5.748 | -0.2354 | Yes | ||
| 82 | TNFAIP3 | 2900142 | 18498 | -6.065 | -0.1689 | Yes | ||
| 83 | CCL5 | 3710397 | 18577 | -7.590 | -0.0882 | Yes | ||
| 84 | CCRK | 1450161 2030500 5080112 | 18584 | -8.059 | 0.0017 | Yes |

