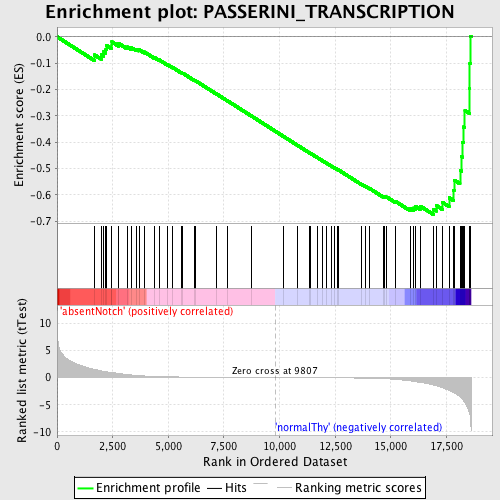
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | PASSERINI_TRANSCRIPTION |
| Enrichment Score (ES) | -0.6747056 |
| Normalized Enrichment Score (NES) | -1.6002264 |
| Nominal p-value | 0.0018903592 |
| FDR q-value | 0.25335082 |
| FWER p-Value | 0.947 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KLF9 | 3840022 | 1682 | 1.518 | -0.0688 | No | ||
| 2 | TP53 | 6130707 | 1986 | 1.229 | -0.0675 | No | ||
| 3 | RRN3 | 5550138 | 2098 | 1.145 | -0.0570 | No | ||
| 4 | TRIM29 | 4560369 5900121 | 2191 | 1.058 | -0.0468 | No | ||
| 5 | ATF4 | 730441 1740195 | 2208 | 1.046 | -0.0326 | No | ||
| 6 | TBX6 | 4730139 6840647 | 2460 | 0.935 | -0.0327 | No | ||
| 7 | NFKB1 | 5420358 | 2462 | 0.934 | -0.0194 | No | ||
| 8 | FUBP1 | 5390373 | 2769 | 0.735 | -0.0253 | No | ||
| 9 | ETS1 | 5270278 6450717 6620465 | 3159 | 0.542 | -0.0385 | No | ||
| 10 | SOX8 | 580026 | 3331 | 0.476 | -0.0409 | No | ||
| 11 | KLF4 | 1850022 3830239 5570750 | 3574 | 0.389 | -0.0483 | No | ||
| 12 | TWIST1 | 5130619 | 3680 | 0.358 | -0.0488 | No | ||
| 13 | EN1 | 5360168 7050025 | 3921 | 0.294 | -0.0575 | No | ||
| 14 | NR2F2 | 3170609 3310577 | 4356 | 0.213 | -0.0778 | No | ||
| 15 | MYOG | 3190672 | 4581 | 0.180 | -0.0873 | No | ||
| 16 | ETV1 | 70735 2940603 5080463 | 4979 | 0.138 | -0.1068 | No | ||
| 17 | HR | 2690095 6200300 | 5198 | 0.120 | -0.1168 | No | ||
| 18 | HOXC10 | 5360170 | 5573 | 0.097 | -0.1355 | No | ||
| 19 | TCF12 | 3610324 7000156 | 5640 | 0.094 | -0.1377 | No | ||
| 20 | CTCF | 5340017 | 6156 | 0.073 | -0.1644 | No | ||
| 21 | FEV | 840369 5860139 | 6207 | 0.071 | -0.1661 | No | ||
| 22 | MEF2C | 670025 780338 | 7167 | 0.044 | -0.2172 | No | ||
| 23 | BRD8 | 1170463 2900075 4050397 6590332 | 7652 | 0.035 | -0.2427 | No | ||
| 24 | HOXD10 | 6100039 | 8735 | 0.016 | -0.3008 | No | ||
| 25 | ENPP2 | 5860546 | 10196 | -0.006 | -0.3794 | No | ||
| 26 | THRB | 1780673 | 10789 | -0.015 | -0.4111 | No | ||
| 27 | TAF4 | 1450170 | 11337 | -0.025 | -0.4402 | No | ||
| 28 | JUN | 840170 | 11408 | -0.026 | -0.4436 | No | ||
| 29 | MITF | 380056 | 11690 | -0.031 | -0.4583 | No | ||
| 30 | DLX4 | 50215 6650059 | 11939 | -0.037 | -0.4711 | No | ||
| 31 | GATA6 | 1770010 | 12104 | -0.041 | -0.4794 | No | ||
| 32 | PITX2 | 870537 2690139 | 12327 | -0.046 | -0.4907 | No | ||
| 33 | HAND2 | 4670142 | 12478 | -0.050 | -0.4981 | No | ||
| 34 | HOXB5 | 2900538 | 12623 | -0.054 | -0.5051 | No | ||
| 35 | MEIS1 | 1400575 | 12643 | -0.054 | -0.5053 | No | ||
| 36 | SPDEF | 360070 | 13694 | -0.098 | -0.5605 | No | ||
| 37 | TEAD3 | 2320142 | 13861 | -0.110 | -0.5678 | No | ||
| 38 | FOS | 1850315 | 14050 | -0.124 | -0.5762 | No | ||
| 39 | RXRB | 1780040 5340438 | 14691 | -0.196 | -0.6079 | No | ||
| 40 | NR2C2 | 6100097 | 14699 | -0.197 | -0.6054 | No | ||
| 41 | ARNT | 1170672 5670711 | 14810 | -0.219 | -0.6082 | No | ||
| 42 | SNAPC1 | 6860324 | 15211 | -0.324 | -0.6251 | No | ||
| 43 | DMTF1 | 5910047 1570497 2480100 6620239 | 15871 | -0.603 | -0.6519 | No | ||
| 44 | KLF5 | 3840348 | 16019 | -0.684 | -0.6500 | No | ||
| 45 | AIP | 5720300 | 16110 | -0.737 | -0.6443 | No | ||
| 46 | ERCC3 | 6900008 | 16321 | -0.891 | -0.6428 | No | ||
| 47 | RARA | 4050161 | 16914 | -1.364 | -0.6551 | Yes | ||
| 48 | NFYA | 5860368 | 17037 | -1.493 | -0.6402 | Yes | ||
| 49 | NFKBIA | 1570152 | 17332 | -1.860 | -0.6294 | Yes | ||
| 50 | NFKB2 | 2320670 | 17622 | -2.349 | -0.6112 | Yes | ||
| 51 | NFE2L1 | 6130053 | 17813 | -2.718 | -0.5824 | Yes | ||
| 52 | E4F1 | 6110717 | 17848 | -2.798 | -0.5440 | Yes | ||
| 53 | STAT1 | 6510204 6590553 | 18110 | -3.562 | -0.5069 | Yes | ||
| 54 | FOXO1A | 3120670 | 18186 | -3.857 | -0.4556 | Yes | ||
| 55 | SMAD3 | 6450671 | 18213 | -3.958 | -0.4001 | Yes | ||
| 56 | STAT3 | 460040 3710341 | 18266 | -4.219 | -0.3423 | Yes | ||
| 57 | GTF3C1 | 1990368 4210037 | 18332 | -4.592 | -0.2798 | Yes | ||
| 58 | PBX2 | 3130433 6510292 | 18544 | -6.601 | -0.1964 | Yes | ||
| 59 | XBP1 | 3840594 | 18551 | -6.754 | -0.0997 | Yes | ||
| 60 | ISGF3G | 3190131 7100142 | 18568 | -7.181 | 0.0026 | Yes |

