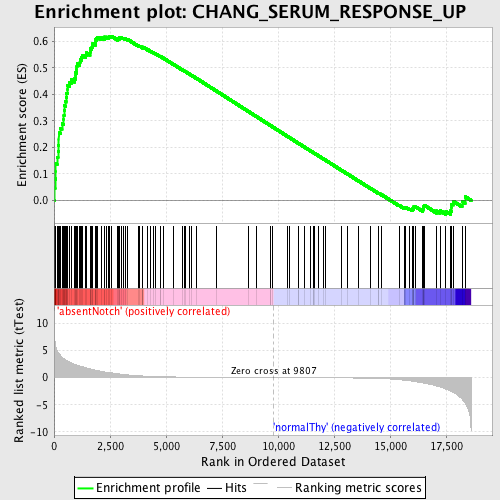
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | CHANG_SERUM_RESPONSE_UP |
| Enrichment Score (ES) | 0.61965966 |
| Normalized Enrichment Score (NES) | 1.6659266 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.037189864 |
| FWER p-Value | 0.527 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPL37 | 3710286 6420088 | 9 | 8.943 | 0.0474 | Yes | ||
| 2 | NOLA2 | 4060167 | 41 | 6.591 | 0.0811 | Yes | ||
| 3 | IFRD2 | 4150110 | 77 | 5.595 | 0.1092 | Yes | ||
| 4 | LYAR | 7000402 | 81 | 5.535 | 0.1387 | Yes | ||
| 5 | JTV1 | 460324 | 133 | 5.018 | 0.1628 | Yes | ||
| 6 | MRPS28 | 2230373 | 175 | 4.681 | 0.1857 | Yes | ||
| 7 | MRPL12 | 3120112 | 204 | 4.467 | 0.2081 | Yes | ||
| 8 | RUVBL1 | 4280239 5080273 | 217 | 4.398 | 0.2311 | Yes | ||
| 9 | PA2G4 | 2030463 | 218 | 4.396 | 0.2546 | Yes | ||
| 10 | PSMD14 | 5690593 | 291 | 4.035 | 0.2724 | Yes | ||
| 11 | IPO4 | 5050619 | 353 | 3.741 | 0.2891 | Yes | ||
| 12 | RFC3 | 1980600 | 432 | 3.495 | 0.3036 | Yes | ||
| 13 | RNASEH2A | 4850270 | 438 | 3.486 | 0.3220 | Yes | ||
| 14 | TCEB1 | 1170619 | 447 | 3.431 | 0.3400 | Yes | ||
| 15 | NME1 | 770014 | 457 | 3.394 | 0.3577 | Yes | ||
| 16 | PSMA7 | 2230131 | 498 | 3.257 | 0.3730 | Yes | ||
| 17 | SDC1 | 3440471 6350408 | 533 | 3.182 | 0.3882 | Yes | ||
| 18 | LMNB2 | 5900692 | 551 | 3.143 | 0.4041 | Yes | ||
| 19 | PSMC3 | 2480184 | 603 | 3.036 | 0.4177 | Yes | ||
| 20 | PAICS | 610142 | 608 | 3.017 | 0.4336 | Yes | ||
| 21 | SRM | 1850097 2480162 | 698 | 2.834 | 0.4440 | Yes | ||
| 22 | NUTF2 | 1410471 5720050 | 761 | 2.707 | 0.4551 | Yes | ||
| 23 | MCM3 | 5570068 | 925 | 2.409 | 0.4592 | Yes | ||
| 24 | IL7R | 2060088 6380500 | 959 | 2.354 | 0.4701 | Yes | ||
| 25 | CCT5 | 1050670 | 965 | 2.341 | 0.4823 | Yes | ||
| 26 | ENO1 | 5340128 | 1009 | 2.287 | 0.4923 | Yes | ||
| 27 | SMS | 2120707 4060162 | 1016 | 2.281 | 0.5042 | Yes | ||
| 28 | NUP107 | 6380021 | 1025 | 2.269 | 0.5159 | Yes | ||
| 29 | PSMD12 | 730044 | 1154 | 2.117 | 0.5203 | Yes | ||
| 30 | LSM4 | 3130180 | 1159 | 2.107 | 0.5314 | Yes | ||
| 31 | UMPS | 2340541 4730438 | 1214 | 2.044 | 0.5394 | Yes | ||
| 32 | TOMM40 | 6110017 | 1256 | 1.984 | 0.5479 | Yes | ||
| 33 | HMGN2 | 3140091 | 1422 | 1.774 | 0.5484 | Yes | ||
| 34 | PDAP1 | 1410735 | 1443 | 1.746 | 0.5567 | Yes | ||
| 35 | TXNL2 | 70563 5700433 5720373 | 1603 | 1.577 | 0.5566 | Yes | ||
| 36 | EIF4G1 | 4070446 | 1608 | 1.575 | 0.5648 | Yes | ||
| 37 | RBMX | 7100162 2900541 | 1619 | 1.566 | 0.5726 | Yes | ||
| 38 | PLAUR | 5910280 | 1687 | 1.512 | 0.5771 | Yes | ||
| 39 | PPIH | 5910403 | 1722 | 1.471 | 0.5832 | Yes | ||
| 40 | MRPS16 | 2510390 | 1729 | 1.465 | 0.5907 | Yes | ||
| 41 | MYBL2 | 5220609 6520368 | 1832 | 1.374 | 0.5925 | Yes | ||
| 42 | SNRPD1 | 4480162 | 1850 | 1.362 | 0.5989 | Yes | ||
| 43 | LSM3 | 780164 | 1854 | 1.358 | 0.6060 | Yes | ||
| 44 | CORO1C | 4570397 | 1886 | 1.320 | 0.6114 | Yes | ||
| 45 | PSMD2 | 4670706 5050364 | 1939 | 1.273 | 0.6155 | Yes | ||
| 46 | TPI1 | 1500215 2100154 | 2093 | 1.147 | 0.6133 | Yes | ||
| 47 | VDAC1 | 4760059 | 2241 | 1.029 | 0.6109 | Yes | ||
| 48 | SFRS2 | 50707 380593 | 2242 | 1.028 | 0.6164 | Yes | ||
| 49 | HMGB1 | 2120670 2350044 | 2342 | 0.997 | 0.6164 | Yes | ||
| 50 | TNFRSF12A | 4810253 | 2443 | 0.943 | 0.6160 | Yes | ||
| 51 | CDK2 | 130484 2260301 4010088 5050110 | 2469 | 0.928 | 0.6197 | Yes | ||
| 52 | MCM7 | 3290292 5220056 | 2567 | 0.868 | 0.6191 | No | ||
| 53 | EEF1E1 | 4540711 | 2850 | 0.695 | 0.6075 | No | ||
| 54 | TPM2 | 520735 3870390 | 2852 | 0.695 | 0.6112 | No | ||
| 55 | SSR3 | 1400040 | 2904 | 0.668 | 0.6120 | No | ||
| 56 | BRIP1 | 1050427 | 2910 | 0.665 | 0.6153 | No | ||
| 57 | TUBG1 | 3140204 | 2999 | 0.621 | 0.6139 | No | ||
| 58 | CBX1 | 5080408 1980239 | 3089 | 0.578 | 0.6122 | No | ||
| 59 | H2AFZ | 1470168 | 3180 | 0.533 | 0.6102 | No | ||
| 60 | EMP2 | 580128 | 3286 | 0.493 | 0.6071 | No | ||
| 61 | SLC25A5 | 1980672 | 3761 | 0.331 | 0.5833 | No | ||
| 62 | CDCA4 | 5890315 6110487 | 3814 | 0.319 | 0.5822 | No | ||
| 63 | SNRPA | 2570100 4150121 6940484 | 3958 | 0.285 | 0.5760 | No | ||
| 64 | SNRPB | 1230097 2940086 3520671 | 3963 | 0.283 | 0.5773 | No | ||
| 65 | BRCA2 | 4280372 | 3965 | 0.282 | 0.5787 | No | ||
| 66 | COTL1 | 3840050 6350215 | 4190 | 0.238 | 0.5679 | No | ||
| 67 | TPM1 | 130673 | 4315 | 0.220 | 0.5623 | No | ||
| 68 | HAS2 | 5360181 | 4428 | 0.201 | 0.5574 | No | ||
| 69 | NLN | 1450576 5870575 | 4515 | 0.189 | 0.5537 | No | ||
| 70 | NUPL1 | 360273 6770100 | 4734 | 0.161 | 0.5428 | No | ||
| 71 | GNG11 | 4010142 | 4741 | 0.160 | 0.5433 | No | ||
| 72 | CENPJ | 6660551 4670592 | 4868 | 0.148 | 0.5373 | No | ||
| 73 | FLNC | 2360048 5050270 | 5307 | 0.113 | 0.5142 | No | ||
| 74 | SLC16A1 | 50433 | 5750 | 0.089 | 0.4908 | No | ||
| 75 | PLG | 3360270 3840100 | 5809 | 0.086 | 0.4881 | No | ||
| 76 | UBE2J1 | 4070133 | 5865 | 0.084 | 0.4856 | No | ||
| 77 | HNRPR | 2320440 2900601 4610008 | 6038 | 0.078 | 0.4767 | No | ||
| 78 | MT3 | 1450537 | 6123 | 0.074 | 0.4725 | No | ||
| 79 | CHEK1 | 50167 1940300 | 6368 | 0.066 | 0.4597 | No | ||
| 80 | SNRPE | 5080021 | 7256 | 0.043 | 0.4120 | No | ||
| 81 | POLE2 | 6110041 | 8698 | 0.017 | 0.3341 | No | ||
| 82 | F3 | 2940180 | 9038 | 0.011 | 0.3158 | No | ||
| 83 | MTHFD1 | 5900398 6520427 | 9650 | 0.003 | 0.2828 | No | ||
| 84 | TFPI2 | 3870324 | 9674 | 0.002 | 0.2816 | No | ||
| 85 | MET | 2690348 | 9734 | 0.001 | 0.2784 | No | ||
| 86 | RBM14 | 5340731 5690315 | 10440 | -0.010 | 0.2403 | No | ||
| 87 | MNAT1 | 730292 2350364 | 10505 | -0.011 | 0.2369 | No | ||
| 88 | MKKS | 2760722 3390563 | 10893 | -0.017 | 0.2160 | No | ||
| 89 | TAGLN | 6660280 | 10931 | -0.017 | 0.2141 | No | ||
| 90 | PNN | 6650504 | 11196 | -0.022 | 0.2000 | No | ||
| 91 | RPN1 | 1240113 | 11446 | -0.027 | 0.1867 | No | ||
| 92 | ADAMTS1 | 3140286 5860463 | 11565 | -0.029 | 0.1804 | No | ||
| 93 | MYCBP | 4280528 | 11567 | -0.029 | 0.1805 | No | ||
| 94 | RNF41 | 1050711 5820333 | 11615 | -0.030 | 0.1781 | No | ||
| 95 | EPHB1 | 4610056 | 11791 | -0.033 | 0.1689 | No | ||
| 96 | MYBL1 | 2900168 | 12014 | -0.038 | 0.1571 | No | ||
| 97 | GGH | 4850373 6290152 | 12107 | -0.041 | 0.1523 | No | ||
| 98 | GPLD1 | 6100121 6290154 | 12847 | -0.061 | 0.1127 | No | ||
| 99 | DHFR | 6350315 | 13114 | -0.070 | 0.0986 | No | ||
| 100 | LOXL2 | 730017 | 13596 | -0.093 | 0.0731 | No | ||
| 101 | COPS6 | 1850195 | 14129 | -0.131 | 0.0450 | No | ||
| 102 | SNRPA1 | 1050735 6110131 | 14482 | -0.165 | 0.0269 | No | ||
| 103 | COX17 | 6250072 | 14499 | -0.167 | 0.0269 | No | ||
| 104 | EBNA1BP2 | 50369 1400433 6040019 | 14634 | -0.189 | 0.0207 | No | ||
| 105 | PLOD2 | 3360427 | 15397 | -0.390 | -0.0184 | No | ||
| 106 | POLR3K | 1400577 5290500 | 15644 | -0.495 | -0.0291 | No | ||
| 107 | MAPRE1 | 3290037 | 15648 | -0.496 | -0.0266 | No | ||
| 108 | NUDT1 | 2850600 | 15681 | -0.512 | -0.0256 | No | ||
| 109 | PITPNC1 | 3990017 | 15873 | -0.604 | -0.0327 | No | ||
| 110 | MAP3K8 | 2940286 | 15987 | -0.666 | -0.0352 | No | ||
| 111 | UAP1 | 3060093 | 15996 | -0.673 | -0.0320 | No | ||
| 112 | PFN1 | 6130132 | 16033 | -0.689 | -0.0303 | No | ||
| 113 | SSSCA1 | 1240402 780309 | 16056 | -0.701 | -0.0277 | No | ||
| 114 | SMURF2 | 1940161 | 16062 | -0.703 | -0.0242 | No | ||
| 115 | MYL6 | 60563 6100152 | 16138 | -0.754 | -0.0242 | No | ||
| 116 | SFRS10 | 6130528 6400438 7040112 | 16451 | -0.976 | -0.0359 | No | ||
| 117 | PFKP | 70138 6760040 1170278 | 16468 | -0.985 | -0.0315 | No | ||
| 118 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 16491 | -0.998 | -0.0273 | No | ||
| 119 | BCCIP | 670450 2650750 5270519 6660711 | 16499 | -1.000 | -0.0223 | No | ||
| 120 | DCLRE1B | 540070 4730373 5550176 | 16531 | -1.021 | -0.0185 | No | ||
| 121 | RNF138 | 110142 3870129 6020441 6400193 | 17075 | -1.543 | -0.0396 | No | ||
| 122 | EIF4EBP1 | 60132 5720148 | 17228 | -1.733 | -0.0386 | No | ||
| 123 | MSN | 4060044 | 17490 | -2.112 | -0.0414 | No | ||
| 124 | PTS | 1500270 | 17708 | -2.515 | -0.0396 | No | ||
| 125 | WSB2 | 2900433 | 17742 | -2.581 | -0.0276 | No | ||
| 126 | ITGA6 | 3830129 | 17748 | -2.591 | -0.0140 | No | ||
| 127 | PCSK7 | 6370450 | 17812 | -2.715 | -0.0028 | No | ||
| 128 | VIL2 | 5570161 | 18211 | -3.944 | -0.0032 | No | ||
| 129 | DCK | 6900411 | 18345 | -4.675 | 0.0147 | No |

