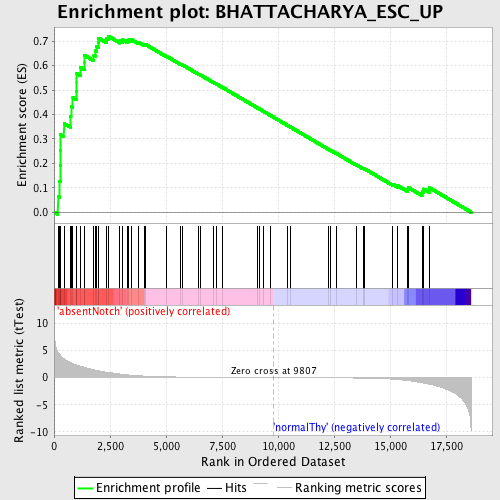
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | BHATTACHARYA_ESC_UP |
| Enrichment Score (ES) | 0.7207574 |
| Normalized Enrichment Score (NES) | 1.7410716 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016986202 |
| FWER p-Value | 0.109 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NME2 | 2190672 | 173 | 4.687 | 0.0648 | Yes | ||
| 2 | PPAT | 1980019 | 255 | 4.206 | 0.1269 | Yes | ||
| 3 | SERPINH1 | 6130014 | 276 | 4.115 | 0.1909 | Yes | ||
| 4 | IMPDH2 | 5220138 | 294 | 4.012 | 0.2535 | Yes | ||
| 5 | DDX21 | 6100446 | 298 | 3.996 | 0.3165 | Yes | ||
| 6 | CCT8 | 6220019 | 441 | 3.449 | 0.3634 | Yes | ||
| 7 | RPS24 | 5420377 | 733 | 2.747 | 0.3912 | Yes | ||
| 8 | SNRPF | 3170280 | 754 | 2.717 | 0.4331 | Yes | ||
| 9 | TNNT1 | 3360671 | 840 | 2.543 | 0.4687 | Yes | ||
| 10 | EIF4A1 | 1990341 2810300 | 1010 | 2.286 | 0.4957 | Yes | ||
| 11 | NASP | 2260139 2940369 5130707 | 1015 | 2.282 | 0.5316 | Yes | ||
| 12 | SMS | 2120707 4060162 | 1016 | 2.281 | 0.5677 | Yes | ||
| 13 | MAD2L2 | 1240358 | 1178 | 2.086 | 0.5920 | Yes | ||
| 14 | KIF4A | 1410465 4760070 | 1344 | 1.888 | 0.6130 | Yes | ||
| 15 | CCNC | 3390750 | 1347 | 1.885 | 0.6427 | Yes | ||
| 16 | GDF3 | 510551 | 1777 | 1.421 | 0.6420 | Yes | ||
| 17 | MTHFD2 | 3800411 | 1845 | 1.368 | 0.6600 | Yes | ||
| 18 | GAL | 3930279 | 1885 | 1.322 | 0.6789 | Yes | ||
| 19 | PSMA2 | 6510093 | 1964 | 1.252 | 0.6945 | Yes | ||
| 20 | ELOVL6 | 5340746 | 1989 | 1.229 | 0.7126 | Yes | ||
| 21 | PSIP1 | 1780082 3190435 5050594 | 2325 | 1.000 | 0.7104 | Yes | ||
| 22 | GJA1 | 5220731 | 2415 | 0.961 | 0.7208 | Yes | ||
| 23 | HDAC2 | 4050433 | 2940 | 0.654 | 0.7029 | No | ||
| 24 | TK1 | 540014 540494 6760152 | 3070 | 0.589 | 0.7052 | No | ||
| 25 | RPL24 | 3870273 | 3274 | 0.497 | 0.7022 | No | ||
| 26 | RPL4 | 2940014 5420324 | 3340 | 0.471 | 0.7061 | No | ||
| 27 | CALU | 2680403 3520056 4610348 5720176 6130121 | 3461 | 0.429 | 0.7064 | No | ||
| 28 | RPL7 | 3840647 | 3766 | 0.330 | 0.6953 | No | ||
| 29 | PODXL | 130717 | 4034 | 0.269 | 0.6851 | No | ||
| 30 | KPNA2 | 3450114 | 4093 | 0.257 | 0.6861 | No | ||
| 31 | RPL6 | 6040735 | 5020 | 0.135 | 0.6383 | No | ||
| 32 | CCNB1 | 4590433 4780372 | 5661 | 0.093 | 0.6053 | No | ||
| 33 | SLC16A1 | 50433 | 5750 | 0.089 | 0.6019 | No | ||
| 34 | SSB | 460286 3140717 | 6460 | 0.063 | 0.5647 | No | ||
| 35 | EPRS | 3170301 | 6554 | 0.060 | 0.5607 | No | ||
| 36 | GSH1 | 2760242 | 7099 | 0.046 | 0.5321 | No | ||
| 37 | CRABP1 | 450278 | 7269 | 0.042 | 0.5237 | No | ||
| 38 | SFRP2 | 4850097 | 7538 | 0.037 | 0.5098 | No | ||
| 39 | HMGB2 | 2640603 | 9069 | 0.011 | 0.4275 | No | ||
| 40 | CRABP2 | 6940725 | 9161 | 0.009 | 0.4228 | No | ||
| 41 | NPM1 | 4730427 | 9337 | 0.007 | 0.4135 | No | ||
| 42 | MTHFD1 | 5900398 6520427 | 9650 | 0.003 | 0.3967 | No | ||
| 43 | SET | 6650286 | 10418 | -0.010 | 0.3555 | No | ||
| 44 | TDGF1 | 1690338 6760070 | 10545 | -0.011 | 0.3489 | No | ||
| 45 | LAPTM4B | 2060377 4150398 | 12233 | -0.044 | 0.2587 | No | ||
| 46 | PITX2 | 870537 2690139 | 12327 | -0.046 | 0.2544 | No | ||
| 47 | FABP5 | 730452 4810497 | 12611 | -0.053 | 0.2400 | No | ||
| 48 | SEMA6A | 2690138 3060021 | 13484 | -0.085 | 0.1944 | No | ||
| 49 | CYP26A1 | 380022 2690600 | 13810 | -0.106 | 0.1786 | No | ||
| 50 | LRRN1 | 3290154 | 13857 | -0.110 | 0.1778 | No | ||
| 51 | DSG2 | 360528 2630603 | 15126 | -0.297 | 0.1142 | No | ||
| 52 | HNRPAB | 540504 | 15345 | -0.371 | 0.1083 | No | ||
| 53 | MGST1 | 6020605 | 15774 | -0.556 | 0.0940 | No | ||
| 54 | PTTG1 | 520463 | 15797 | -0.567 | 0.1018 | No | ||
| 55 | HSPA4 | 1050170 3450309 | 16426 | -0.960 | 0.0832 | No | ||
| 56 | PSMA3 | 5900047 7040161 | 16501 | -1.000 | 0.0950 | No | ||
| 57 | IDH1 | 1990021 | 16747 | -1.195 | 0.1007 | No |

