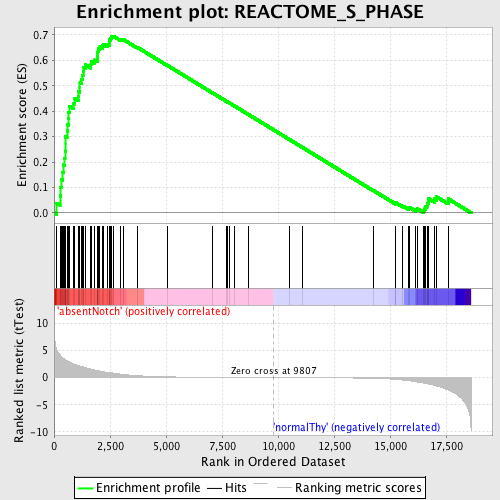
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | REACTOME_S_PHASE |
| Enrichment Score (ES) | 0.69553226 |
| Normalized Enrichment Score (NES) | 1.7378237 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022681147 |
| FWER p-Value | 0.057 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB1 | 2940402 | 118 | 5.165 | 0.0387 | Yes | ||
| 2 | POLD2 | 6400148 | 265 | 4.153 | 0.0670 | Yes | ||
| 3 | PSMD14 | 5690593 | 291 | 4.035 | 0.1008 | Yes | ||
| 4 | PSMA4 | 4560427 | 339 | 3.803 | 0.1314 | Yes | ||
| 5 | CCNH | 3190347 | 389 | 3.617 | 0.1603 | Yes | ||
| 6 | RFC3 | 1980600 | 432 | 3.495 | 0.1885 | Yes | ||
| 7 | PSMC5 | 2760315 6550021 | 469 | 3.356 | 0.2158 | Yes | ||
| 8 | PSME3 | 2810537 | 487 | 3.284 | 0.2435 | Yes | ||
| 9 | RFC1 | 1190673 | 497 | 3.258 | 0.2714 | Yes | ||
| 10 | PSMA7 | 2230131 | 498 | 3.257 | 0.2998 | Yes | ||
| 11 | RFC4 | 3800082 6840142 | 578 | 3.094 | 0.3225 | Yes | ||
| 12 | PSMC3 | 2480184 | 603 | 3.036 | 0.3477 | Yes | ||
| 13 | PSMD8 | 630142 | 645 | 2.912 | 0.3709 | Yes | ||
| 14 | LIG1 | 1980438 4540112 5570609 | 654 | 2.897 | 0.3957 | Yes | ||
| 15 | CUL1 | 1990632 | 678 | 2.868 | 0.4195 | Yes | ||
| 16 | PSMD3 | 1400647 | 849 | 2.522 | 0.4323 | Yes | ||
| 17 | MCM3 | 5570068 | 925 | 2.409 | 0.4492 | Yes | ||
| 18 | PRIM1 | 6420746 | 1091 | 2.192 | 0.4594 | Yes | ||
| 19 | MCM4 | 2760673 5420711 | 1104 | 2.179 | 0.4778 | Yes | ||
| 20 | PSMB3 | 4280594 | 1152 | 2.119 | 0.4937 | Yes | ||
| 21 | PSMD12 | 730044 | 1154 | 2.117 | 0.5121 | Yes | ||
| 22 | PSMB6 | 2690711 | 1234 | 2.020 | 0.5255 | Yes | ||
| 23 | CDKN1A | 4050088 6400706 | 1270 | 1.970 | 0.5407 | Yes | ||
| 24 | PSMC4 | 580050 1580025 | 1299 | 1.941 | 0.5561 | Yes | ||
| 25 | MCM6 | 60092 540181 6510110 | 1326 | 1.907 | 0.5714 | Yes | ||
| 26 | PSMC2 | 7040010 | 1411 | 1.790 | 0.5824 | Yes | ||
| 27 | PSMB5 | 6290242 | 1644 | 1.542 | 0.5834 | Yes | ||
| 28 | POLA2 | 940519 | 1657 | 1.535 | 0.5961 | Yes | ||
| 29 | CDC25A | 3800184 | 1784 | 1.416 | 0.6016 | Yes | ||
| 30 | PSMC1 | 6350538 | 1930 | 1.286 | 0.6050 | Yes | ||
| 31 | PSMD2 | 4670706 5050364 | 1939 | 1.273 | 0.6157 | Yes | ||
| 32 | POLE | 6020538 | 1952 | 1.262 | 0.6261 | Yes | ||
| 33 | PSMC6 | 2810021 | 1957 | 1.260 | 0.6368 | Yes | ||
| 34 | PSMA2 | 6510093 | 1964 | 1.252 | 0.6474 | Yes | ||
| 35 | MCM5 | 2680647 | 2047 | 1.184 | 0.6533 | Yes | ||
| 36 | PSMB10 | 630504 6510039 | 2146 | 1.098 | 0.6576 | Yes | ||
| 37 | RFC5 | 3800452 6020091 | 2217 | 1.040 | 0.6629 | Yes | ||
| 38 | MCM2 | 5050139 | 2398 | 0.971 | 0.6616 | Yes | ||
| 39 | ORC5L | 1940133 1940711 | 2458 | 0.936 | 0.6666 | Yes | ||
| 40 | CDK2 | 130484 2260301 4010088 5050110 | 2469 | 0.928 | 0.6742 | Yes | ||
| 41 | CCND1 | 460524 770309 3120576 6980398 | 2475 | 0.923 | 0.6819 | Yes | ||
| 42 | RPA2 | 2760301 5420195 | 2529 | 0.885 | 0.6868 | Yes | ||
| 43 | MCM7 | 3290292 5220056 | 2567 | 0.868 | 0.6924 | Yes | ||
| 44 | PSMA5 | 5390537 | 2641 | 0.816 | 0.6955 | Yes | ||
| 45 | POLD1 | 4830026 | 2960 | 0.641 | 0.6840 | No | ||
| 46 | PSMD7 | 2030619 6220594 | 3079 | 0.581 | 0.6827 | No | ||
| 47 | ORC2L | 1990470 6510019 | 3710 | 0.349 | 0.6517 | No | ||
| 48 | PSMA6 | 50609 | 5043 | 0.133 | 0.5811 | No | ||
| 49 | POLA1 | 4760541 | 7071 | 0.046 | 0.4722 | No | ||
| 50 | WEE1 | 3390070 | 7693 | 0.034 | 0.4390 | No | ||
| 51 | CDK7 | 2640451 | 7747 | 0.033 | 0.4364 | No | ||
| 52 | ORC4L | 4230538 5550288 | 7842 | 0.031 | 0.4316 | No | ||
| 53 | PSMA1 | 380059 2760195 | 8066 | 0.027 | 0.4198 | No | ||
| 54 | POLE2 | 6110041 | 8698 | 0.017 | 0.3859 | No | ||
| 55 | MNAT1 | 730292 2350364 | 10505 | -0.011 | 0.2886 | No | ||
| 56 | PSMD5 | 3940139 4570041 | 11087 | -0.020 | 0.2575 | No | ||
| 57 | PSMB9 | 6980471 | 14256 | -0.143 | 0.0879 | No | ||
| 58 | PSMD11 | 2340538 6510053 | 15242 | -0.337 | 0.0377 | No | ||
| 59 | CDC45L | 70537 3130114 | 15261 | -0.345 | 0.0397 | No | ||
| 60 | POLD3 | 6400278 | 15571 | -0.461 | 0.0271 | No | ||
| 61 | CDK4 | 540075 4540600 | 15811 | -0.575 | 0.0192 | No | ||
| 62 | PSMB4 | 520402 | 15868 | -0.602 | 0.0214 | No | ||
| 63 | RPA3 | 5700136 | 16147 | -0.757 | 0.0130 | No | ||
| 64 | PCNA | 940754 | 16217 | -0.813 | 0.0164 | No | ||
| 65 | PSMA3 | 5900047 7040161 | 16501 | -1.000 | 0.0098 | No | ||
| 66 | RPA1 | 360452 | 16553 | -1.031 | 0.0161 | No | ||
| 67 | PSMD9 | 50020 5890368 | 16581 | -1.046 | 0.0237 | No | ||
| 68 | FEN1 | 1770541 | 16647 | -1.102 | 0.0298 | No | ||
| 69 | PSMD4 | 430068 | 16674 | -1.128 | 0.0383 | No | ||
| 70 | PSME1 | 450193 4480035 | 16709 | -1.157 | 0.0465 | No | ||
| 71 | ORC1L | 2370328 6110390 | 16727 | -1.176 | 0.0558 | No | ||
| 72 | RB1 | 5900338 | 16977 | -1.425 | 0.0548 | No | ||
| 73 | PSMD10 | 520494 1170576 3830050 | 17066 | -1.530 | 0.0634 | No | ||
| 74 | PSMB2 | 940035 4210324 | 17591 | -2.307 | 0.0553 | No |

