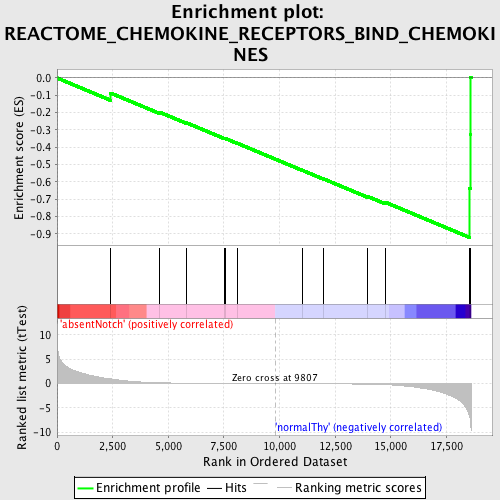
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES |
| Enrichment Score (ES) | -0.9213498 |
| Normalized Enrichment Score (NES) | -1.6801053 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.043131467 |
| FWER p-Value | 0.273 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCL25 | 450541 540435 | 2401 | 0.970 | -0.0870 | No | ||
| 2 | IL8RB | 450592 1170537 | 4610 | 0.176 | -0.1980 | No | ||
| 3 | CXCL11 | 1090551 | 5803 | 0.086 | -0.2583 | No | ||
| 4 | XCR1 | 5080072 | 7517 | 0.037 | -0.3488 | No | ||
| 5 | CCR3 | 50427 | 7561 | 0.037 | -0.3495 | No | ||
| 6 | CCR10 | 4050097 | 8128 | 0.026 | -0.3788 | No | ||
| 7 | CX3CL1 | 3990707 | 11023 | -0.019 | -0.5335 | No | ||
| 8 | IL8RA | 1580100 | 11957 | -0.037 | -0.5821 | No | ||
| 9 | CXCR6 | 3190440 | 13966 | -0.118 | -0.6849 | No | ||
| 10 | CCR6 | 5720368 6020176 | 14770 | -0.211 | -0.7189 | Yes | ||
| 11 | XCL1 | 3800504 | 18537 | -6.532 | -0.6379 | Yes | ||
| 12 | CCR7 | 2060687 | 18569 | -7.207 | -0.3269 | Yes | ||
| 13 | CCL5 | 3710397 | 18577 | -7.590 | 0.0021 | Yes |

