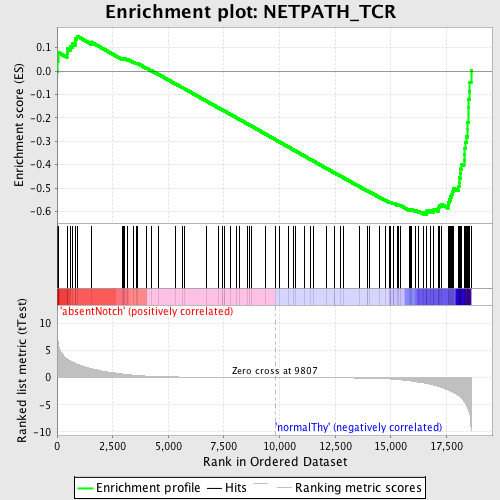
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.61311364 |
| Normalized Enrichment Score (NES) | -1.566161 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13590229 |
| FWER p-Value | 0.93 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 1780148 | 20 | 7.873 | 0.0435 | No | ||
| 2 | CEBPB | 2970019 | 40 | 6.620 | 0.0800 | No | ||
| 3 | UNC119 | 360372 | 446 | 3.435 | 0.0776 | No | ||
| 4 | GRB2 | 6650398 | 481 | 3.303 | 0.0945 | No | ||
| 5 | PTPN22 | 1690647 7100239 | 585 | 3.076 | 0.1064 | No | ||
| 6 | STAT5A | 2680458 | 695 | 2.835 | 0.1165 | No | ||
| 7 | PIK3R2 | 1660193 | 807 | 2.604 | 0.1253 | No | ||
| 8 | YWHAQ | 6760524 | 834 | 2.555 | 0.1384 | No | ||
| 9 | NCL | 2360463 4540279 | 893 | 2.461 | 0.1492 | No | ||
| 10 | PTPRC | 130402 5290148 | 1532 | 1.647 | 0.1241 | No | ||
| 11 | HOMER3 | 3830279 | 2943 | 0.651 | 0.0516 | No | ||
| 12 | ABL1 | 1050593 2030050 4010114 | 2989 | 0.624 | 0.0527 | No | ||
| 13 | CREBBP | 5690035 7040050 | 3008 | 0.616 | 0.0552 | No | ||
| 14 | ARHGEF6 | 4780164 6380450 | 3184 | 0.532 | 0.0488 | No | ||
| 15 | SHB | 4920494 | 3449 | 0.433 | 0.0370 | No | ||
| 16 | RAPGEF1 | 60040 4590670 | 3559 | 0.395 | 0.0333 | No | ||
| 17 | STK39 | 50692 | 3615 | 0.379 | 0.0325 | No | ||
| 18 | DUSP3 | 1240129 6840215 | 4025 | 0.271 | 0.0119 | No | ||
| 19 | MAPK1 | 3190193 6200253 | 4254 | 0.228 | 0.0009 | No | ||
| 20 | CDC42 | 1240168 3440278 4480519 5290162 | 4538 | 0.186 | -0.0133 | No | ||
| 21 | ENAH | 1690292 5700300 | 5326 | 0.112 | -0.0552 | No | ||
| 22 | PLCG1 | 6020369 | 5644 | 0.094 | -0.0718 | No | ||
| 23 | PAG1 | 5910397 | 5703 | 0.091 | -0.0744 | No | ||
| 24 | GRAP2 | 7100441 1410647 | 6704 | 0.056 | -0.1281 | No | ||
| 25 | PTPN11 | 2230100 2470180 6100528 | 7253 | 0.043 | -0.1575 | No | ||
| 26 | LYN | 6040600 | 7419 | 0.039 | -0.1662 | No | ||
| 27 | PIK3R1 | 4730671 | 7432 | 0.039 | -0.1666 | No | ||
| 28 | PTPRJ | 4010707 | 7521 | 0.037 | -0.1711 | No | ||
| 29 | INSL3 | 4150092 | 7529 | 0.037 | -0.1713 | No | ||
| 30 | AKT1 | 5290746 | 7793 | 0.032 | -0.1853 | No | ||
| 31 | MAP2K1 | 840739 | 8058 | 0.027 | -0.1994 | No | ||
| 32 | RAP1A | 1090025 | 8179 | 0.025 | -0.2058 | No | ||
| 33 | CREB1 | 1500717 2230358 3610600 6550601 | 8536 | 0.020 | -0.2249 | No | ||
| 34 | TUBB2A | 6940435 | 8661 | 0.018 | -0.2315 | No | ||
| 35 | TXK | 2320338 | 8747 | 0.016 | -0.2360 | No | ||
| 36 | PTPRH | 3870070 | 9344 | 0.007 | -0.2682 | No | ||
| 37 | NFATC2 | 70450 540097 | 9799 | 0.000 | -0.2927 | No | ||
| 38 | SKAP2 | 6760500 | 9994 | -0.003 | -0.3031 | No | ||
| 39 | PTPN3 | 870369 | 10413 | -0.009 | -0.3257 | No | ||
| 40 | PRKCQ | 2260170 3870193 | 10603 | -0.012 | -0.3358 | No | ||
| 41 | GAB2 | 1410280 2340520 4280040 | 10728 | -0.014 | -0.3424 | No | ||
| 42 | SOS1 | 7050338 | 11112 | -0.021 | -0.3630 | No | ||
| 43 | JUN | 840170 | 11408 | -0.026 | -0.3788 | No | ||
| 44 | CD2AP | 1940369 | 11502 | -0.028 | -0.3837 | No | ||
| 45 | ARHGEF7 | 1690441 | 12123 | -0.041 | -0.4169 | No | ||
| 46 | NCK1 | 6200575 6510050 | 12465 | -0.049 | -0.4350 | No | ||
| 47 | SRC | 580132 | 12487 | -0.050 | -0.4359 | No | ||
| 48 | ACP1 | 1990435 | 12738 | -0.057 | -0.4491 | No | ||
| 49 | SYK | 6940133 | 12894 | -0.062 | -0.4571 | No | ||
| 50 | SLA | 2000070 | 13587 | -0.092 | -0.4940 | No | ||
| 51 | SHC1 | 2900731 3170504 6520537 | 13933 | -0.115 | -0.5119 | No | ||
| 52 | DLG1 | 2630091 6020286 | 13942 | -0.116 | -0.5117 | No | ||
| 53 | FOS | 1850315 | 14050 | -0.124 | -0.5168 | No | ||
| 54 | DBNL | 7100088 | 14494 | -0.166 | -0.5398 | No | ||
| 55 | PTPN12 | 2030309 6020725 6130746 6290609 | 14741 | -0.205 | -0.5519 | No | ||
| 56 | FYB | 1690440 | 14935 | -0.245 | -0.5609 | No | ||
| 57 | CBLB | 3520465 4670348 | 14969 | -0.256 | -0.5613 | No | ||
| 58 | STAT5B | 6200026 | 14990 | -0.263 | -0.5609 | No | ||
| 59 | SOS2 | 2060722 | 15106 | -0.293 | -0.5654 | No | ||
| 60 | JAK3 | 70347 3290008 | 15141 | -0.301 | -0.5655 | No | ||
| 61 | RIPK2 | 5050072 6290632 | 15296 | -0.354 | -0.5719 | No | ||
| 62 | KHDRBS1 | 1240403 6040040 | 15347 | -0.371 | -0.5725 | No | ||
| 63 | SH2D3C | 3060014 | 15426 | -0.401 | -0.5744 | No | ||
| 64 | VAV3 | 1050731 2450242 2680653 | 15836 | -0.584 | -0.5932 | No | ||
| 65 | FYN | 2100468 4760520 4850687 | 15899 | -0.618 | -0.5930 | No | ||
| 66 | ABI1 | 2850152 | 15943 | -0.636 | -0.5917 | No | ||
| 67 | ZAP70 | 1410494 2260504 | 16100 | -0.731 | -0.5960 | No | ||
| 68 | LCP2 | 2680066 6650707 | 16233 | -0.826 | -0.5985 | No | ||
| 69 | VASP | 7050500 | 16479 | -0.992 | -0.6061 | No | ||
| 70 | SH3BP2 | 610131 | 16610 | -1.072 | -0.6070 | Yes | ||
| 71 | WIPF1 | 1170577 | 16615 | -1.078 | -0.6011 | Yes | ||
| 72 | CRK | 1230162 4780128 | 16625 | -1.085 | -0.5955 | Yes | ||
| 73 | MAP4K1 | 4060692 | 16760 | -1.214 | -0.5958 | Yes | ||
| 74 | GRAP | 1770195 | 16915 | -1.366 | -0.5964 | Yes | ||
| 75 | DNM2 | 50176 3170279 | 16937 | -1.388 | -0.5897 | Yes | ||
| 76 | ARHGDIB | 4730433 | 17124 | -1.603 | -0.5907 | Yes | ||
| 77 | LAT | 3170025 | 17151 | -1.641 | -0.5828 | Yes | ||
| 78 | RASA1 | 1240315 | 17172 | -1.670 | -0.5744 | Yes | ||
| 79 | PTPN6 | 4230128 | 17289 | -1.812 | -0.5704 | Yes | ||
| 80 | TUBA4A | 6900279 | 17572 | -2.264 | -0.5728 | Yes | ||
| 81 | DOCK2 | 2940040 | 17584 | -2.287 | -0.5604 | Yes | ||
| 82 | GIT2 | 6040039 6770181 | 17645 | -2.385 | -0.5501 | Yes | ||
| 83 | CBL | 6380068 | 17677 | -2.437 | -0.5380 | Yes | ||
| 84 | DTX1 | 5900372 | 17731 | -2.550 | -0.5264 | Yes | ||
| 85 | LCK | 3360142 | 17786 | -2.667 | -0.5142 | Yes | ||
| 86 | WAS | 5270193 | 17806 | -2.699 | -0.5000 | Yes | ||
| 87 | WASF2 | 4850592 | 18043 | -3.321 | -0.4939 | Yes | ||
| 88 | EVL | 1740113 | 18072 | -3.430 | -0.4760 | Yes | ||
| 89 | BCL10 | 2360397 | 18080 | -3.458 | -0.4567 | Yes | ||
| 90 | NEDD9 | 1740373 4230053 | 18125 | -3.600 | -0.4387 | Yes | ||
| 91 | VAV1 | 6020487 | 18137 | -3.653 | -0.4186 | Yes | ||
| 92 | CRKL | 4050427 | 18162 | -3.756 | -0.3986 | Yes | ||
| 93 | CD4 | 1090010 | 18290 | -4.362 | -0.3808 | Yes | ||
| 94 | CD3D | 2810739 | 18306 | -4.485 | -0.3562 | Yes | ||
| 95 | CISH | 840315 | 18331 | -4.586 | -0.3315 | Yes | ||
| 96 | CARD11 | 70338 | 18363 | -4.788 | -0.3060 | Yes | ||
| 97 | PSTPIP1 | 3190156 | 18388 | -4.968 | -0.2792 | Yes | ||
| 98 | SH2D2A | 2970594 | 18446 | -5.512 | -0.2510 | Yes | ||
| 99 | CD3E | 3800056 | 18448 | -5.536 | -0.2197 | Yes | ||
| 100 | CD247 | 3800725 5720136 | 18488 | -5.985 | -0.1879 | Yes | ||
| 101 | DEF6 | 840593 | 18489 | -6.000 | -0.1539 | Yes | ||
| 102 | ITK | 2230592 | 18492 | -6.023 | -0.1199 | Yes | ||
| 103 | PTK2B | 4730411 | 18527 | -6.405 | -0.0854 | Yes | ||
| 104 | CD3G | 2680288 | 18554 | -6.817 | -0.0482 | Yes | ||
| 105 | CD5 | 6590338 | 18606 | -9.083 | 0.0005 | Yes |

