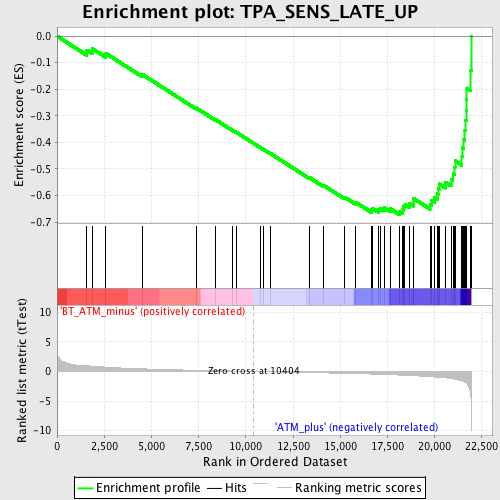
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | TPA_SENS_LATE_UP |
| Enrichment Score (ES) | -0.67152095 |
| Normalized Enrichment Score (NES) | -2.2735589 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.614749E-4 |
| FWER p-Value | 0.0020 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NRP1 | 1418084_at 1448943_at 1448944_at 1457198_at | 1577 | 0.953 | -0.0524 | No | ||
| 2 | CLK3 | 1423849_a_at 1441472_at | 1853 | 0.889 | -0.0466 | No | ||
| 3 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 2585 | 0.723 | -0.0651 | No | ||
| 4 | SERPINA6 | 1448506_at | 4522 | 0.438 | -0.1445 | No | ||
| 5 | IMPDH1 | 1423239_at | 7366 | 0.197 | -0.2703 | No | ||
| 6 | MYLK | 1425504_at 1425505_at 1425506_at | 8406 | 0.126 | -0.3152 | No | ||
| 7 | VIL1 | 1448837_at | 9282 | 0.071 | -0.3537 | No | ||
| 8 | AQP4 | 1425382_a_at 1434449_at 1447745_at | 9513 | 0.057 | -0.3630 | No | ||
| 9 | IL1RAP | 1421843_at 1439697_at 1442614_at 1449585_at | 10783 | -0.023 | -0.4205 | No | ||
| 10 | EXT1 | 1417730_at 1439877_at 1440011_at 1440092_at 1456655_at 1458054_at 1458296_at | 10938 | -0.035 | -0.4268 | No | ||
| 11 | DCT | 1418028_at | 11290 | -0.056 | -0.4417 | No | ||
| 12 | S100A11 | 1460351_at | 13356 | -0.184 | -0.5323 | No | ||
| 13 | SLC13A3 | 1416560_at 1438377_x_at 1439384_at 1439385_x_at | 14121 | -0.236 | -0.5623 | No | ||
| 14 | MMP15 | 1422597_at 1437462_x_at 1439734_at | 15224 | -0.316 | -0.6061 | No | ||
| 15 | CCT5 | 1417258_at | 15795 | -0.359 | -0.6248 | No | ||
| 16 | IL1RN | 1423017_a_at 1425663_at 1451798_at | 16631 | -0.432 | -0.6540 | No | ||
| 17 | SERPINE1 | 1419149_at | 16723 | -0.439 | -0.6491 | No | ||
| 18 | HBEGF | 1418349_at 1418350_at | 17029 | -0.472 | -0.6533 | No | ||
| 19 | AGA | 1434665_at | 17133 | -0.482 | -0.6481 | No | ||
| 20 | RFXAP | 1418859_at | 17328 | -0.502 | -0.6466 | No | ||
| 21 | ALOX5AP | 1452016_at 1459234_at | 17637 | -0.537 | -0.6495 | No | ||
| 22 | ALAS1 | 1424126_at 1442331_at 1455282_x_at | 18119 | -0.600 | -0.6591 | Yes | ||
| 23 | RAC2 | 1417620_at 1440208_at | 18302 | -0.623 | -0.6546 | Yes | ||
| 24 | CDC37 | 1416819_at 1416820_at | 18323 | -0.626 | -0.6426 | Yes | ||
| 25 | ALDH3B2 | 1456317_at | 18423 | -0.640 | -0.6339 | Yes | ||
| 26 | EMP3 | 1417104_at | 18681 | -0.681 | -0.6316 | Yes | ||
| 27 | EVI2A | 1443894_at 1450241_a_at 1458788_at 1460055_at | 18860 | -0.707 | -0.6251 | Yes | ||
| 28 | CYBA | 1454268_a_at | 18861 | -0.707 | -0.6105 | Yes | ||
| 29 | DPP4 | 1416697_at 1438279_at 1441242_at 1441342_at 1459973_x_at | 19750 | -0.873 | -0.6331 | Yes | ||
| 30 | P2RY5 | 1428615_at 1444627_at | 19829 | -0.892 | -0.6182 | Yes | ||
| 31 | DYNLL1 | 1448682_at | 19983 | -0.929 | -0.6060 | Yes | ||
| 32 | PLAUR | 1452521_a_at | 20150 | -0.969 | -0.5936 | Yes | ||
| 33 | KCNN3 | 1421632_at 1459308_at | 20187 | -0.977 | -0.5751 | Yes | ||
| 34 | RARG | 1419415_a_at 1419416_a_at | 20230 | -0.992 | -0.5566 | Yes | ||
| 35 | PREP | 1422796_at 1436198_at 1439942_at | 20566 | -1.066 | -0.5498 | Yes | ||
| 36 | NKRF | 1434398_at 1441325_at 1445983_at | 20866 | -1.187 | -0.5390 | Yes | ||
| 37 | SPRY2 | 1421656_at 1436584_at | 20966 | -1.241 | -0.5179 | Yes | ||
| 38 | CAP1 | 1417461_at 1417462_at | 21047 | -1.281 | -0.4951 | Yes | ||
| 39 | INHBA | 1422053_at 1458291_at | 21074 | -1.299 | -0.4695 | Yes | ||
| 40 | VIM | 1438118_x_at 1450641_at 1456292_a_at | 21441 | -1.597 | -0.4533 | Yes | ||
| 41 | IL2RG | 1416295_a_at 1416296_at | 21451 | -1.604 | -0.4206 | Yes | ||
| 42 | RPS6KA1 | 1416896_at | 21546 | -1.713 | -0.3895 | Yes | ||
| 43 | IFNGR2 | 1423557_at 1423558_at | 21599 | -1.814 | -0.3544 | Yes | ||
| 44 | TSC22D1 | 1425742_a_at 1433899_x_at 1447360_at 1454758_a_at 1454971_x_at 1456132_x_at | 21613 | -1.847 | -0.3169 | Yes | ||
| 45 | ANXA5 | 1425567_a_at | 21661 | -1.941 | -0.2790 | Yes | ||
| 46 | FLNA | 1426677_at | 21670 | -1.984 | -0.2384 | Yes | ||
| 47 | TRAF1 | 1423602_at 1445452_at | 21704 | -2.081 | -0.1970 | Yes | ||
| 48 | DUSP6 | 1415834_at | 21907 | -3.749 | -0.1288 | Yes | ||
| 49 | EGR2 | 1427682_a_at 1427683_at | 21930 | -6.298 | 0.0002 | Yes |

