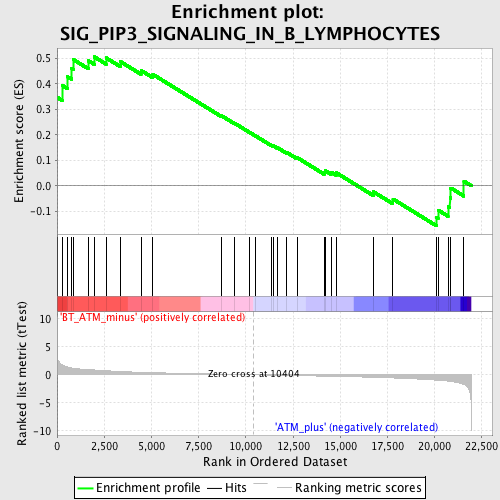
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES |
| Enrichment Score (ES) | 0.5058863 |
| Normalized Enrichment Score (NES) | 1.6517876 |
| Nominal p-value | 0.021531101 |
| FDR q-value | 0.28201473 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP1R13B | 1434895_s_at 1442956_at 1459224_at | 0 | 10.211 | 0.3470 | Yes | ||
| 2 | PTPRC | 1422124_a_at 1440165_at | 268 | 1.734 | 0.3938 | Yes | ||
| 3 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 557 | 1.383 | 0.4276 | Yes | ||
| 4 | FLOT2 | 1417544_a_at 1436666_at 1438164_x_at | 765 | 1.230 | 0.4600 | Yes | ||
| 5 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 854 | 1.165 | 0.4955 | Yes | ||
| 6 | ITPR2 | 1421678_at 1424833_at 1424834_s_at 1427287_s_at 1427693_at 1444418_at 1444551_at 1447328_at | 1656 | 0.935 | 0.4908 | Yes | ||
| 7 | PDK1 | 1423747_a_at 1423748_at 1434974_at 1435836_at | 1969 | 0.864 | 0.5059 | Yes | ||
| 8 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 2595 | 0.722 | 0.5019 | No | ||
| 9 | FOXO3A | 1434831_a_at 1434832_at 1439598_at 1444226_at 1450236_at 1458279_at | 3353 | 0.586 | 0.4872 | No | ||
| 10 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 4445 | 0.446 | 0.4526 | No | ||
| 11 | RPS6KB1 | 1428849_at 1446376_at 1454956_at 1457562_at 1459951_at 1460705_at | 5073 | 0.384 | 0.4370 | No | ||
| 12 | NR0B2 | 1449854_at | 8712 | 0.107 | 0.2745 | No | ||
| 13 | FLOT1 | 1448559_at | 9387 | 0.065 | 0.2460 | No | ||
| 14 | PITX2 | 1424797_a_at 1450482_a_at | 10173 | 0.014 | 0.2106 | No | ||
| 15 | BTK | 1422755_at | 10520 | -0.006 | 0.1950 | No | ||
| 16 | TEC | 1441524_at 1460204_at | 11357 | -0.059 | 0.1589 | No | ||
| 17 | CDKN2A | 1450140_a_at | 11446 | -0.065 | 0.1571 | No | ||
| 18 | SAG | 1419025_at | 11651 | -0.078 | 0.1504 | No | ||
| 19 | RPS6KA2 | 1417542_at 1417543_at 1441311_at | 12172 | -0.109 | 0.1303 | No | ||
| 20 | GAB1 | 1417693_a_at 1417694_at 1448814_at | 12712 | -0.143 | 0.1106 | No | ||
| 21 | AKT3 | 1422078_at 1435260_at 1435879_at 1443222_at 1460307_at | 14140 | -0.237 | 0.0535 | No | ||
| 22 | DAPP1 | 1421936_at 1421937_at 1447337_at | 14193 | -0.241 | 0.0593 | No | ||
| 23 | PTEN | 1422553_at 1441593_at 1444325_at 1450655_at 1454722_at 1455728_at 1457493_at | 14509 | -0.263 | 0.0539 | No | ||
| 24 | BCR | 1427265_at 1452368_at 1455564_at | 14774 | -0.284 | 0.0515 | No | ||
| 25 | CD19 | 1450570_a_at | 16753 | -0.443 | -0.0238 | No | ||
| 26 | PLCG2 | 1426926_at | 17787 | -0.555 | -0.0521 | No | ||
| 27 | VAV1 | 1422932_a_at | 20103 | -0.957 | -0.1253 | No | ||
| 28 | PSCD3 | 1418758_a_at 1431707_a_at 1446268_at | 20177 | -0.975 | -0.0955 | No | ||
| 29 | AKT2 | 1421324_a_at 1424480_s_at 1455703_at | 20705 | -1.113 | -0.0817 | No | ||
| 30 | RPS6KA3 | 1427299_at 1441360_at 1452383_at | 20807 | -1.159 | -0.0469 | No | ||
| 31 | ITPR3 | 1417297_at | 20851 | -1.179 | -0.0088 | No | ||
| 32 | RPS6KA1 | 1416896_at | 21546 | -1.713 | 0.0177 | No |

