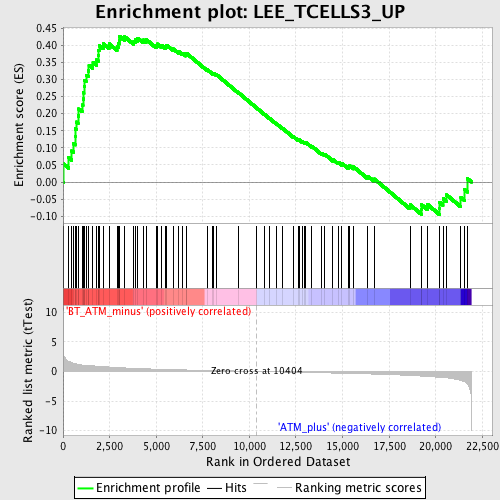
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | LEE_TCELLS3_UP |
| Enrichment Score (ES) | 0.42631608 |
| Normalized Enrichment Score (NES) | 1.7219294 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.45053285 |
| FWER p-Value | 0.982 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DIAPH3 | 1422944_a_at 1441628_at 1443445_at 1445803_at 1459374_at | 25 | 2.963 | 0.0532 | Yes | ||
| 2 | TTK | 1442594_at 1449171_at | 303 | 1.668 | 0.0711 | Yes | ||
| 3 | MPP1 | 1423243_at 1450919_at | 468 | 1.490 | 0.0910 | Yes | ||
| 4 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 557 | 1.383 | 0.1123 | Yes | ||
| 5 | KIF11 | 1435306_a_at 1452314_at 1452315_at | 646 | 1.306 | 0.1322 | Yes | ||
| 6 | CEP70 | 1422653_at 1430231_a_at | 656 | 1.299 | 0.1557 | Yes | ||
| 7 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 725 | 1.254 | 0.1756 | Yes | ||
| 8 | NEIL3 | 1424060_at 1440633_at | 805 | 1.197 | 0.1939 | Yes | ||
| 9 | BUB1 | 1424046_at 1438571_at | 844 | 1.170 | 0.2136 | Yes | ||
| 10 | SLC1A4 | 1423549_at 1423550_at 1456003_a_at | 1022 | 1.083 | 0.2254 | Yes | ||
| 11 | CCNB1 | 1419943_s_at 1419944_at | 1069 | 1.069 | 0.2429 | Yes | ||
| 12 | RAD51AP1 | 1417938_at 1417939_at 1417940_s_at 1448899_s_at | 1089 | 1.064 | 0.2616 | Yes | ||
| 13 | GALNT7 | 1425581_s_at 1426908_at 1445598_at 1452232_at | 1122 | 1.052 | 0.2794 | Yes | ||
| 14 | NUSAP1 | 1416309_at 1416310_at 1442830_at | 1137 | 1.048 | 0.2980 | Yes | ||
| 15 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 1236 | 1.023 | 0.3123 | Yes | ||
| 16 | DTL | 1430228_at 1434695_at 1443864_at 1453753_at 1456652_at | 1351 | 0.999 | 0.3254 | Yes | ||
| 17 | CENPA | 1441864_x_at 1444416_at 1450842_a_at | 1389 | 0.993 | 0.3419 | Yes | ||
| 18 | GGH | 1437522_x_at 1456595_x_at 1460310_a_at 1460613_x_at | 1603 | 0.947 | 0.3495 | Yes | ||
| 19 | CDCA2 | 1437251_at 1455983_at 1459254_at | 1804 | 0.901 | 0.3569 | Yes | ||
| 20 | DLG7 | 1438811_at 1455730_at | 1885 | 0.882 | 0.3694 | Yes | ||
| 21 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 1904 | 0.878 | 0.3847 | Yes | ||
| 22 | DNTT | 1420166_at 1449757_x_at 1450545_a_at | 1942 | 0.869 | 0.3990 | Yes | ||
| 23 | CDKN3 | 1415968_a_at 1415969_s_at 1430574_at | 2153 | 0.821 | 0.4044 | Yes | ||
| 24 | CD99 | 1428850_x_at 1430514_a_at 1453556_x_at | 2468 | 0.750 | 0.4038 | Yes | ||
| 25 | PRC1 | 1423774_a_at 1423775_s_at | 2896 | 0.665 | 0.3965 | Yes | ||
| 26 | MLF1IP | 1428518_at 1452816_at | 2964 | 0.654 | 0.4054 | Yes | ||
| 27 | CDCA1 | 1430811_a_at | 3011 | 0.643 | 0.4151 | Yes | ||
| 28 | OIP5 | 1430617_at | 3024 | 0.642 | 0.4263 | Yes | ||
| 29 | UBE2C | 1452954_at | 3308 | 0.593 | 0.4243 | No | ||
| 30 | TFDP2 | 1435344_at 1437174_at 1443962_at 1445873_at | 3798 | 0.522 | 0.4115 | No | ||
| 31 | PON1 | 1418190_at | 3882 | 0.512 | 0.4170 | No | ||
| 32 | WHSC1 | 1428827_at 1435136_at 1436036_at 1455228_at | 4007 | 0.498 | 0.4205 | No | ||
| 33 | HMMR | 1425815_a_at 1427541_x_at 1429871_at 1450156_a_at 1450157_a_at | 4310 | 0.461 | 0.4152 | No | ||
| 34 | MCM4 | 1416214_at 1436708_x_at | 4455 | 0.445 | 0.4167 | No | ||
| 35 | E2F2 | 1455790_at 1458574_at | 5002 | 0.391 | 0.3989 | No | ||
| 36 | KNTC2 | 1417445_at | 5058 | 0.385 | 0.4035 | No | ||
| 37 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 5294 | 0.363 | 0.3994 | No | ||
| 38 | MCM2 | 1434079_s_at 1448777_at | 5509 | 0.344 | 0.3959 | No | ||
| 39 | HMGB3 | 1416155_at | 5551 | 0.340 | 0.4002 | No | ||
| 40 | HELLS | 1417541_at 1430139_at 1453361_at | 5922 | 0.306 | 0.3889 | No | ||
| 41 | CCNB2 | 1450920_at | 6215 | 0.283 | 0.3807 | No | ||
| 42 | CEP55 | 1452242_at 1453683_a_at | 6414 | 0.268 | 0.3766 | No | ||
| 43 | E2F8 | 1436186_at 1444319_at 1448080_at | 6599 | 0.253 | 0.3728 | No | ||
| 44 | CDCA3 | 1448000_at 1448001_x_at 1452040_a_at | 6628 | 0.251 | 0.3761 | No | ||
| 45 | KIF15 | 1431718_at | 7744 | 0.172 | 0.3283 | No | ||
| 46 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 8038 | 0.151 | 0.3176 | No | ||
| 47 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 8093 | 0.147 | 0.3179 | No | ||
| 48 | SCRN1 | 1428717_at 1428718_at | 8218 | 0.138 | 0.3147 | No | ||
| 49 | UBE2T | 1422462_at 1442398_at | 9423 | 0.063 | 0.2608 | No | ||
| 50 | CKS2 | 1417457_at 1417458_s_at | 10376 | 0.002 | 0.2172 | No | ||
| 51 | MELK | 1416558_at | 10812 | -0.026 | 0.1978 | No | ||
| 52 | RAPGEF5 | 1445255_at 1455137_at 1455840_at | 11097 | -0.044 | 0.1856 | No | ||
| 53 | BIRC5 | 1424278_a_at | 11464 | -0.066 | 0.1701 | No | ||
| 54 | DSCR1 | 1416600_a_at 1416601_a_at | 11789 | -0.086 | 0.1568 | No | ||
| 55 | ASPM | 1422814_at 1441520_at 1452459_at 1458560_at 1458873_at | 12345 | -0.121 | 0.1336 | No | ||
| 56 | KIF20A | 1449207_a_at | 12637 | -0.138 | 0.1229 | No | ||
| 57 | CPVL | 1432056_at | 12673 | -0.140 | 0.1238 | No | ||
| 58 | SGOL2 | 1437370_at | 12840 | -0.152 | 0.1190 | No | ||
| 59 | ATAD2 | 1436174_at 1443146_at 1443229_at | 12958 | -0.158 | 0.1165 | No | ||
| 60 | KIF14 | 1445525_at | 13015 | -0.162 | 0.1170 | No | ||
| 61 | GFI1 | 1417679_at | 13343 | -0.183 | 0.1053 | No | ||
| 62 | AURKA | 1424511_at 1440129_at | 13890 | -0.221 | 0.0844 | No | ||
| 63 | TUBB | 1416256_a_at 1455719_at | 14053 | -0.232 | 0.0812 | No | ||
| 64 | TSHR | 1421999_at 1428001_at | 14490 | -0.261 | 0.0661 | No | ||
| 65 | PBK | 1448627_s_at | 14772 | -0.284 | 0.0584 | No | ||
| 66 | PALLD | 1427228_at 1433768_at 1440635_at 1440693_at | 14970 | -0.298 | 0.0549 | No | ||
| 67 | PPIL5 | 1427506_at 1436922_at 1452458_s_at | 15331 | -0.324 | 0.0444 | No | ||
| 68 | ADA | 1417976_at | 15395 | -0.329 | 0.0475 | No | ||
| 69 | UHRF1 | 1415810_at 1415811_at 1439227_at | 15599 | -0.343 | 0.0445 | No | ||
| 70 | CXXC5 | 1431469_a_at 1448960_at | 16350 | -0.405 | 0.0176 | No | ||
| 71 | AEBP1 | 1422514_at 1450637_a_at | 16712 | -0.438 | 0.0092 | No | ||
| 72 | TNFRSF21 | 1422740_at 1450731_s_at | 18630 | -0.671 | -0.0663 | No | ||
| 73 | GNA15 | 1421302_a_at | 19235 | -0.772 | -0.0797 | No | ||
| 74 | E2F7 | 1437187_at | 19250 | -0.776 | -0.0661 | No | ||
| 75 | KIF2C | 1432392_at 1437611_x_at 1454221_a_at | 19559 | -0.832 | -0.0650 | No | ||
| 76 | TUBB2C | 1423642_at 1438622_x_at 1439416_x_at 1456031_at 1456078_x_at 1456470_x_at | 20208 | -0.983 | -0.0766 | No | ||
| 77 | LETM1 | 1420825_at 1420826_at 1460284_at | 20219 | -0.988 | -0.0589 | No | ||
| 78 | NEK2 | 1417299_at 1437579_at 1437580_s_at 1443999_at | 20401 | -1.025 | -0.0484 | No | ||
| 79 | RAG1 | 1440574_at 1450680_at | 20591 | -1.074 | -0.0373 | No | ||
| 80 | FAIM | 1418029_at | 21360 | -1.514 | -0.0447 | No | ||
| 81 | CDC25A | 1417131_at 1417132_at 1445995_at | 21559 | -1.732 | -0.0220 | No | ||
| 82 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 21716 | -2.132 | 0.0100 | No |

