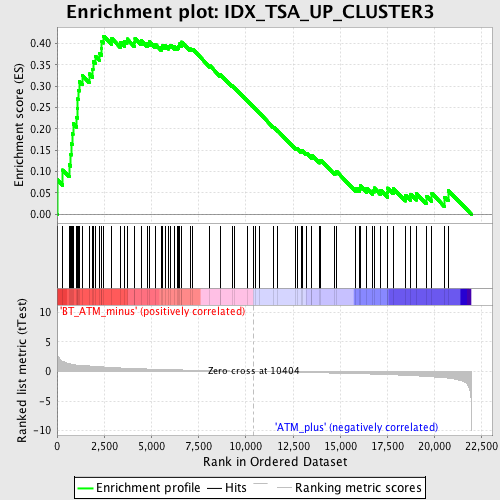
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | IDX_TSA_UP_CLUSTER3 |
| Enrichment Score (ES) | 0.4173271 |
| Normalized Enrichment Score (NES) | 1.6886057 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.3596874 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CENPK | 1418264_at 1437068_at 1457001_at | 10 | 3.797 | 0.0813 | Yes | ||
| 2 | TTK | 1442594_at 1449171_at | 303 | 1.668 | 0.1038 | Yes | ||
| 3 | KIF11 | 1435306_a_at 1452314_at 1452315_at | 646 | 1.306 | 0.1163 | Yes | ||
| 4 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 725 | 1.254 | 0.1397 | Yes | ||
| 5 | NCAPH | 1423920_at 1436707_x_at | 745 | 1.242 | 0.1655 | Yes | ||
| 6 | CCNE2 | 1422535_at | 801 | 1.200 | 0.1888 | Yes | ||
| 7 | BUB1 | 1424046_at 1438571_at | 844 | 1.170 | 0.2121 | Yes | ||
| 8 | USP1 | 1423674_at 1423675_at 1451080_at | 1021 | 1.083 | 0.2274 | Yes | ||
| 9 | CCNB1 | 1419943_s_at 1419944_at | 1069 | 1.069 | 0.2482 | Yes | ||
| 10 | RAD51AP1 | 1417938_at 1417939_at 1417940_s_at 1448899_s_at | 1089 | 1.064 | 0.2703 | Yes | ||
| 11 | NUSAP1 | 1416309_at 1416310_at 1442830_at | 1137 | 1.048 | 0.2907 | Yes | ||
| 12 | KIF4A | 1450692_at | 1181 | 1.036 | 0.3110 | Yes | ||
| 13 | DTL | 1430228_at 1434695_at 1443864_at 1453753_at 1456652_at | 1351 | 0.999 | 0.3248 | Yes | ||
| 14 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 1699 | 0.925 | 0.3288 | Yes | ||
| 15 | DLG7 | 1438811_at 1455730_at | 1885 | 0.882 | 0.3393 | Yes | ||
| 16 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 1904 | 0.878 | 0.3574 | Yes | ||
| 17 | TOPBP1 | 1452241_at 1458480_at | 2016 | 0.851 | 0.3706 | Yes | ||
| 18 | MAN2A1 | 1448647_at | 2246 | 0.799 | 0.3773 | Yes | ||
| 19 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 2356 | 0.772 | 0.3890 | Yes | ||
| 20 | AQP1 | 1416203_at | 2369 | 0.770 | 0.4050 | Yes | ||
| 21 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 2454 | 0.753 | 0.4173 | Yes | ||
| 22 | PRC1 | 1423774_a_at 1423775_s_at | 2896 | 0.665 | 0.4115 | No | ||
| 23 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 3361 | 0.584 | 0.4028 | No | ||
| 24 | MCM5 | 1415945_at 1436808_x_at | 3579 | 0.552 | 0.4048 | No | ||
| 25 | GAS6 | 1417399_at 1459585_at | 3702 | 0.536 | 0.4107 | No | ||
| 26 | SMC2 | 1429658_a_at 1429659_at 1429660_s_at 1448635_at 1458479_at | 4111 | 0.483 | 0.4024 | No | ||
| 27 | MCM3 | 1420029_at 1426653_at | 4122 | 0.482 | 0.4123 | No | ||
| 28 | MCM4 | 1416214_at 1436708_x_at | 4455 | 0.445 | 0.4067 | No | ||
| 29 | ABCA1 | 1421839_at 1421840_at 1450392_at | 4772 | 0.413 | 0.4011 | No | ||
| 30 | FIGNL1 | 1422430_at 1460746_at | 4890 | 0.402 | 0.4044 | No | ||
| 31 | PLK1 | 1443408_at 1448191_at 1459616_at | 5199 | 0.371 | 0.3983 | No | ||
| 32 | MCM2 | 1434079_s_at 1448777_at | 5509 | 0.344 | 0.3916 | No | ||
| 33 | KIF22 | 1423813_at 1437716_x_at 1451128_s_at | 5599 | 0.336 | 0.3948 | No | ||
| 34 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 5751 | 0.322 | 0.3948 | No | ||
| 35 | HELLS | 1417541_at 1430139_at 1453361_at | 5922 | 0.306 | 0.3936 | No | ||
| 36 | RFC5 | 1452917_at | 6006 | 0.299 | 0.3962 | No | ||
| 37 | CCNB2 | 1450920_at | 6215 | 0.283 | 0.3928 | No | ||
| 38 | MAD2L1 | 1422460_at | 6367 | 0.271 | 0.3917 | No | ||
| 39 | DCK | 1428838_a_at 1439012_a_at 1449176_a_at | 6446 | 0.266 | 0.3939 | No | ||
| 40 | DTYMK | 1438096_a_at 1449116_a_at 1452681_at | 6458 | 0.264 | 0.3991 | No | ||
| 41 | FEN1 | 1421731_a_at 1436454_x_at | 6596 | 0.253 | 0.3982 | No | ||
| 42 | E2F8 | 1436186_at 1444319_at 1448080_at | 6599 | 0.253 | 0.4036 | No | ||
| 43 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 7057 | 0.218 | 0.3874 | No | ||
| 44 | SLBP | 1430058_at 1460168_at | 7163 | 0.212 | 0.3871 | No | ||
| 45 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 8093 | 0.147 | 0.3478 | No | ||
| 46 | CCNF | 1422513_at 1443807_x_at | 8633 | 0.112 | 0.3255 | No | ||
| 47 | RRM1 | 1415878_at 1440073_at 1448127_at | 8646 | 0.111 | 0.3274 | No | ||
| 48 | IL13RA1 | 1425625_at 1427164_at 1427165_at 1451775_s_at 1454783_at | 9262 | 0.073 | 0.3008 | No | ||
| 49 | HGF | 1425379_at 1442884_at 1451866_a_at | 9381 | 0.065 | 0.2968 | No | ||
| 50 | OSMR | 1418674_at 1418675_at 1459217_at | 10106 | 0.018 | 0.2640 | No | ||
| 51 | CKS2 | 1417457_at 1417458_s_at | 10376 | 0.002 | 0.2518 | No | ||
| 52 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 10480 | -0.004 | 0.2472 | No | ||
| 53 | ASF1B | 1423714_at | 10712 | -0.018 | 0.2370 | No | ||
| 54 | BIRC5 | 1424278_a_at | 11464 | -0.066 | 0.2040 | No | ||
| 55 | PTGFR | 1420349_at 1440777_x_at 1446331_at 1449828_at 1453924_a_at | 11683 | -0.080 | 0.1958 | No | ||
| 56 | KIF20A | 1449207_a_at | 12637 | -0.138 | 0.1551 | No | ||
| 57 | WNT4 | 1450782_at | 12722 | -0.144 | 0.1544 | No | ||
| 58 | IMPDH2 | 1415851_a_at 1415852_at | 12929 | -0.157 | 0.1483 | No | ||
| 59 | CDC45L | 1416575_at 1457838_at | 12997 | -0.161 | 0.1487 | No | ||
| 60 | BZW1 | 1423040_at 1423041_a_at 1450845_a_at 1450846_at | 13199 | -0.174 | 0.1433 | No | ||
| 61 | NUP85 | 1416073_a_at 1437520_a_at | 13481 | -0.193 | 0.1346 | No | ||
| 62 | RAD51 | 1418281_at | 13482 | -0.193 | 0.1387 | No | ||
| 63 | AURKA | 1424511_at 1440129_at | 13890 | -0.221 | 0.1248 | No | ||
| 64 | SLCO3A1 | 1418030_at 1434537_at 1448918_at | 13960 | -0.225 | 0.1265 | No | ||
| 65 | TUBB6 | 1416431_at 1446353_at | 14696 | -0.278 | 0.0989 | No | ||
| 66 | BRCA1 | 1424629_at 1424630_a_at 1427698_at 1443763_at 1451417_at | 14796 | -0.286 | 0.1005 | No | ||
| 67 | LMO4 | 1420981_a_at 1439651_at | 15827 | -0.362 | 0.0612 | No | ||
| 68 | TCF19 | 1423809_at 1443795_at | 16004 | -0.377 | 0.0612 | No | ||
| 69 | TRIP13 | 1429294_at 1429295_s_at 1446760_at | 16053 | -0.381 | 0.0672 | No | ||
| 70 | CDCA8 | 1428480_at 1428481_s_at 1436847_s_at | 16400 | -0.411 | 0.0603 | No | ||
| 71 | INCENP | 1423092_at 1423093_at 1439436_x_at 1441314_at | 16728 | -0.440 | 0.0548 | No | ||
| 72 | CSRP2 | 1420731_a_at | 16786 | -0.446 | 0.0617 | No | ||
| 73 | ILF2 | 1417948_s_at 1417949_at | 17140 | -0.483 | 0.0560 | No | ||
| 74 | CDC6 | 1417019_a_at | 17494 | -0.521 | 0.0510 | No | ||
| 75 | CDCA7 | 1428069_at 1445681_at | 17510 | -0.522 | 0.0616 | No | ||
| 76 | KLF5 | 1430255_at 1451021_a_at 1451739_at | 17802 | -0.557 | 0.0603 | No | ||
| 77 | NQO1 | 1423627_at | 18441 | -0.643 | 0.0449 | No | ||
| 78 | CSTF2 | 1419644_at 1419645_at 1455523_at | 18731 | -0.689 | 0.0465 | No | ||
| 79 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 19021 | -0.733 | 0.0491 | No | ||
| 80 | KIF2C | 1432392_at 1437611_x_at 1454221_a_at | 19559 | -0.832 | 0.0424 | No | ||
| 81 | TACC3 | 1417450_a_at 1436872_at 1455834_x_at | 19848 | -0.896 | 0.0485 | No | ||
| 82 | ADAM8 | 1416871_at | 20521 | -1.054 | 0.0405 | No | ||
| 83 | EZH2 | 1416544_at 1444263_at | 20722 | -1.122 | 0.0555 | No |

