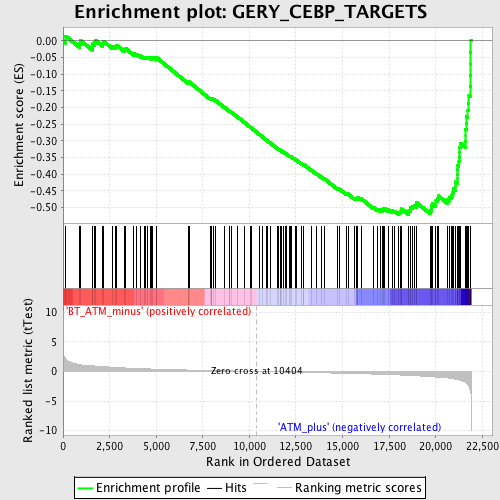
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | GERY_CEBP_TARGETS |
| Enrichment Score (ES) | -0.5205578 |
| Normalized Enrichment Score (NES) | -2.0814352 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0037501808 |
| FWER p-Value | 0.035 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANGPTL4 | 1417130_s_at 1453410_at | 146 | 2.083 | 0.0142 | No | ||
| 2 | PRNP | 1416130_at 1446507_at 1448233_at | 870 | 1.158 | -0.0074 | No | ||
| 3 | DCN | 1441506_at 1449368_at | 936 | 1.127 | 0.0009 | No | ||
| 4 | SERPINB2 | 1419082_at | 1558 | 0.957 | -0.0180 | No | ||
| 5 | NRP1 | 1418084_at 1448943_at 1448944_at 1457198_at | 1577 | 0.953 | -0.0093 | No | ||
| 6 | NSDHL | 1416222_at 1451799_at 1458831_at | 1705 | 0.924 | -0.0059 | No | ||
| 7 | RGS2 | 1419247_at 1419248_at 1447830_s_at | 1755 | 0.912 | 0.0010 | No | ||
| 8 | TOM1L1 | 1439337_at 1451117_a_at | 2137 | 0.825 | -0.0082 | No | ||
| 9 | TNFAIP6 | 1418424_at | 2151 | 0.822 | -0.0006 | No | ||
| 10 | D10ERTD438E | 1419914_s_at 1419915_at 1420646_at 1434304_s_at 1440478_at 1460563_at | 2665 | 0.706 | -0.0171 | No | ||
| 11 | CHORDC1 | 1435574_at 1460645_at | 2791 | 0.683 | -0.0160 | No | ||
| 12 | GREM2 | 1418492_at | 2871 | 0.670 | -0.0129 | No | ||
| 13 | TMEM45A | 1422587_at | 3278 | 0.598 | -0.0255 | No | ||
| 14 | ACSL1 | 1422526_at 1423883_at 1447355_at 1450643_s_at 1460316_at | 3355 | 0.586 | -0.0231 | No | ||
| 15 | IFRD1 | 1416067_at 1442014_at | 3785 | 0.523 | -0.0376 | No | ||
| 16 | CXCL1 | 1419209_at 1441855_x_at 1457644_s_at | 3952 | 0.504 | -0.0401 | No | ||
| 17 | PIM1 | 1423006_at 1435458_at | 4153 | 0.479 | -0.0445 | No | ||
| 18 | LPL | 1415904_at | 4352 | 0.457 | -0.0490 | No | ||
| 19 | GCH1 | 1420499_at 1429692_s_at | 4442 | 0.446 | -0.0487 | No | ||
| 20 | HMOX1 | 1448239_at | 4537 | 0.436 | -0.0486 | No | ||
| 21 | SLFN4 | 1427102_at | 4672 | 0.424 | -0.0505 | No | ||
| 22 | ABCA1 | 1421839_at 1421840_at 1450392_at | 4772 | 0.413 | -0.0509 | No | ||
| 23 | AW112037 | 1435326_at 1435327_at | 4824 | 0.408 | -0.0492 | No | ||
| 24 | C1QDC1 | 1427944_at | 5007 | 0.390 | -0.0536 | No | ||
| 25 | NR2F6 | 1449846_at 1460647_a_at 1460648_at | 5031 | 0.388 | -0.0508 | No | ||
| 26 | KNG1 | 1416676_at 1426045_at | 6729 | 0.244 | -0.1262 | No | ||
| 27 | DNAJB9 | 1417191_at | 6735 | 0.244 | -0.1240 | No | ||
| 28 | BTG2 | 1416250_at 1448272_at | 6769 | 0.241 | -0.1231 | No | ||
| 29 | ADSSL1 | 1449383_at | 7892 | 0.161 | -0.1729 | No | ||
| 30 | GLA | 1418248_at 1449006_at | 7959 | 0.157 | -0.1744 | No | ||
| 31 | IGF2 | 1415931_at 1448152_at | 7973 | 0.156 | -0.1734 | No | ||
| 32 | NXPH2 | 1437964_at | 7993 | 0.155 | -0.1727 | No | ||
| 33 | CXCL5 | 1419728_at | 8070 | 0.149 | -0.1747 | No | ||
| 34 | PPARG | 1420715_a_at | 8208 | 0.139 | -0.1796 | No | ||
| 35 | AI594671 | 1449603_at | 8661 | 0.109 | -0.1993 | No | ||
| 36 | FOXG1 | 1418357_at | 8943 | 0.093 | -0.2112 | No | ||
| 37 | KLF4 | 1417394_at 1417395_at | 8957 | 0.092 | -0.2109 | No | ||
| 38 | FGF7 | 1422243_at 1438405_at | 9040 | 0.087 | -0.2138 | No | ||
| 39 | HGF | 1425379_at 1442884_at 1451866_a_at | 9381 | 0.065 | -0.2287 | No | ||
| 40 | NR1D1 | 1421802_at 1426464_at | 9713 | 0.045 | -0.2435 | No | ||
| 41 | ABCD2 | 1419748_at 1438431_at 1439835_x_at 1446962_at 1456812_at | 10061 | 0.021 | -0.2592 | No | ||
| 42 | FOXF2 | 1418220_at 1447562_at | 10108 | 0.018 | -0.2611 | No | ||
| 43 | HSPA1B | 1427126_at 1427127_x_at 1452318_a_at | 10557 | -0.009 | -0.2816 | No | ||
| 44 | LSS | 1420013_s_at 1420014_at | 10728 | -0.019 | -0.2892 | No | ||
| 45 | ABHD5 | 1417565_at 1417566_at 1447548_at | 10920 | -0.034 | -0.2976 | No | ||
| 46 | RGS16 | 1426037_a_at 1451452_a_at 1455265_a_at | 10953 | -0.035 | -0.2987 | No | ||
| 47 | FHL2 | 1419184_a_at | 11115 | -0.045 | -0.3057 | No | ||
| 48 | TGM3 | 1421355_at 1440150_at | 11511 | -0.069 | -0.3231 | No | ||
| 49 | 2900054D09RIK | 1452872_at | 11557 | -0.072 | -0.3244 | No | ||
| 50 | HSP110 | 1423566_a_at 1425993_a_at | 11652 | -0.078 | -0.3280 | No | ||
| 51 | HSPA1A | 1452388_at | 11655 | -0.078 | -0.3273 | No | ||
| 52 | HTATIP2 | 1451814_a_at 1454227_at | 11712 | -0.082 | -0.3290 | No | ||
| 53 | APBB1IP | 1425269_at 1443145_at | 11814 | -0.088 | -0.3328 | No | ||
| 54 | EREG | 1419431_at | 11930 | -0.095 | -0.3371 | No | ||
| 55 | DDIT3 | 1417516_at 1443897_at | 12017 | -0.099 | -0.3401 | No | ||
| 56 | RASL11B | 1423854_a_at | 12157 | -0.108 | -0.3454 | No | ||
| 57 | HMGCR | 1427229_at 1451766_at | 12209 | -0.111 | -0.3466 | No | ||
| 58 | SH3GL3 | 1432103_a_at | 12281 | -0.115 | -0.3487 | No | ||
| 59 | LOC636537 | 1448843_at | 12471 | -0.128 | -0.3561 | No | ||
| 60 | XDH | 1451006_at | 12517 | -0.131 | -0.3568 | No | ||
| 61 | ZFAND2A | 1415940_at 1415941_s_at 1449702_at 1449703_at | 12813 | -0.150 | -0.3689 | No | ||
| 62 | RBED1 | 1436695_x_at 1451392_at | 12899 | -0.155 | -0.3712 | No | ||
| 63 | IFI204 /// LOC672547 | 1419603_at | 12917 | -0.156 | -0.3704 | No | ||
| 64 | MKNK2 | 1418300_a_at 1449029_at | 13322 | -0.182 | -0.3871 | No | ||
| 65 | CLU | 1418626_a_at 1437458_x_at 1437689_x_at 1454849_x_at | 13588 | -0.200 | -0.3973 | No | ||
| 66 | POSTN | 1423606_at | 13863 | -0.219 | -0.4077 | No | ||
| 67 | TCTE3 | 1421682_a_at 1421683_at | 14016 | -0.229 | -0.4123 | No | ||
| 68 | EIF1AY | 1419736_a_at 1437071_at 1449576_at | 14733 | -0.281 | -0.4424 | No | ||
| 69 | CRABP2 | 1451191_at | 14835 | -0.288 | -0.4441 | No | ||
| 70 | ORM1 | 1451054_at | 15213 | -0.316 | -0.4583 | No | ||
| 71 | CD47 | 1419554_at 1428187_at 1430997_at 1449507_a_at 1458634_at 1459908_at | 15298 | -0.322 | -0.4589 | No | ||
| 72 | ACTA2 | 1416454_s_at 1444105_at 1456658_at | 15642 | -0.347 | -0.4711 | No | ||
| 73 | PLAT | 1415806_at 1458553_at | 15763 | -0.356 | -0.4731 | No | ||
| 74 | ELOVL6 | 1417403_at 1417404_at 1445062_at 1445578_at 1445641_at | 15815 | -0.361 | -0.4718 | No | ||
| 75 | MVD | 1417303_at 1448663_s_at | 15818 | -0.361 | -0.4683 | No | ||
| 76 | DNAJB10 | 1448657_a_at | 16017 | -0.378 | -0.4736 | No | ||
| 77 | S100A3 | 1421856_at | 16654 | -0.434 | -0.4984 | No | ||
| 78 | TGOLN1 /// TGOLN2 | 1423307_s_at | 16894 | -0.457 | -0.5048 | No | ||
| 79 | HBEGF | 1418349_at 1418350_at | 17029 | -0.472 | -0.5062 | No | ||
| 80 | ECHS1 | 1452341_at | 17145 | -0.483 | -0.5067 | No | ||
| 81 | PTX3 | 1418666_at 1456903_at | 17192 | -0.488 | -0.5039 | No | ||
| 82 | BAG3 | 1422452_at | 17250 | -0.494 | -0.5016 | No | ||
| 83 | PEA15 | 1416406_at 1416407_at 1459733_at | 17464 | -0.518 | -0.5061 | No | ||
| 84 | GIPC2 | 1417178_at 1457330_at | 17663 | -0.540 | -0.5098 | No | ||
| 85 | CYB5R1 | 1424048_a_at | 17799 | -0.556 | -0.5104 | No | ||
| 86 | U90926 | 1460667_at | 17999 | -0.583 | -0.5137 | No | ||
| 87 | RDH11 | 1418760_at 1449209_a_at | 18097 | -0.596 | -0.5122 | No | ||
| 88 | MAFK | 1418616_at | 18179 | -0.607 | -0.5099 | No | ||
| 89 | CD68 | 1449164_at | 18184 | -0.608 | -0.5040 | No | ||
| 90 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 18547 | -0.660 | -0.5140 | Yes | ||
| 91 | CRYAB | 1416455_a_at 1434369_a_at 1435837_at | 18566 | -0.663 | -0.5081 | Yes | ||
| 92 | D8ERTD354E | 1450868_at | 18635 | -0.672 | -0.5045 | Yes | ||
| 93 | NGFB | 1419675_at | 18653 | -0.675 | -0.4986 | Yes | ||
| 94 | MYD116 | 1448325_at | 18773 | -0.695 | -0.4971 | Yes | ||
| 95 | ATF3 | 1449363_at | 18862 | -0.707 | -0.4940 | Yes | ||
| 96 | SAA3P | 1450826_a_at | 18968 | -0.725 | -0.4916 | Yes | ||
| 97 | DNAJB1 | 1416755_at 1416756_at | 18969 | -0.725 | -0.4843 | Yes | ||
| 98 | USP36 | 1420773_at 1455237_at 1456094_at 1458311_at | 19709 | -0.863 | -0.5096 | Yes | ||
| 99 | DBP | 1418174_at 1438211_s_at | 19762 | -0.876 | -0.5032 | Yes | ||
| 100 | HP | 1448881_at | 19769 | -0.878 | -0.4947 | Yes | ||
| 101 | HIST1H2BC | 1418072_at | 19832 | -0.892 | -0.4886 | Yes | ||
| 102 | ALDH3A1 | 1418752_at | 20021 | -0.938 | -0.4878 | Yes | ||
| 103 | WWC1 | 1420007_at 1420008_s_at 1420009_at 1427261_at 1445080_at | 20022 | -0.938 | -0.4785 | Yes | ||
| 104 | BCKDHB | 1427153_at | 20122 | -0.961 | -0.4734 | Yes | ||
| 105 | PLAUR | 1452521_a_at | 20150 | -0.969 | -0.4649 | Yes | ||
| 106 | TGTP | 1449009_at | 20623 | -1.085 | -0.4757 | Yes | ||
| 107 | RNH1 | 1451201_s_at | 20726 | -1.123 | -0.4691 | Yes | ||
| 108 | CD1D1 | 1449130_at 1449131_s_at | 20833 | -1.170 | -0.4623 | Yes | ||
| 109 | PER2 | 1417602_at 1417603_at 1445892_at 1457350_at | 20928 | -1.221 | -0.4544 | Yes | ||
| 110 | HIST2H3C1 | 1442051_at | 20972 | -1.243 | -0.4439 | Yes | ||
| 111 | BAIAP2 | 1425656_a_at 1435128_at 1451027_at 1451028_at | 21081 | -1.302 | -0.4358 | Yes | ||
| 112 | GLRX | 1416592_at 1416593_at | 21087 | -1.304 | -0.4230 | Yes | ||
| 113 | TRIB3 | 1426065_a_at 1456225_x_at | 21165 | -1.347 | -0.4131 | Yes | ||
| 114 | DUSP1 | 1448830_at | 21168 | -1.349 | -0.3996 | Yes | ||
| 115 | GADD45G | 1453851_a_at | 21192 | -1.365 | -0.3870 | Yes | ||
| 116 | 1100001G20RIK | 1434484_at | 21199 | -1.372 | -0.3736 | Yes | ||
| 117 | PREI4 | 1429144_at 1429639_at 1437953_at 1458701_at | 21256 | -1.427 | -0.3619 | Yes | ||
| 118 | ID3 | 1416630_at | 21267 | -1.433 | -0.3480 | Yes | ||
| 119 | DFNA5H | 1417903_at 1421534_at | 21279 | -1.445 | -0.3340 | Yes | ||
| 120 | SERPINI1 | 1416702_at 1443756_at 1448443_at | 21304 | -1.462 | -0.3205 | Yes | ||
| 121 | LCN2 | 1427747_a_at | 21319 | -1.472 | -0.3064 | Yes | ||
| 122 | S100A8 | 1419394_s_at | 21598 | -1.810 | -0.3010 | Yes | ||
| 123 | HIPK1 | 1421298_a_at 1424540_at 1457515_at | 21619 | -1.856 | -0.2834 | Yes | ||
| 124 | SAT1 | 1420502_at | 21621 | -1.860 | -0.2648 | Yes | ||
| 125 | SLFN2 | 1450165_at | 21657 | -1.938 | -0.2470 | Yes | ||
| 126 | JUNB | 1415899_at | 21672 | -1.992 | -0.2277 | Yes | ||
| 127 | C3 | 1423954_at | 21730 | -2.182 | -0.2085 | Yes | ||
| 128 | PROCR | 1420664_s_at | 21748 | -2.247 | -0.1868 | Yes | ||
| 129 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21794 | -2.532 | -0.1635 | Yes | ||
| 130 | LYZS | 1423547_at | 21855 | -3.078 | -0.1354 | Yes | ||
| 131 | IER2 | 1416442_at | 21874 | -3.262 | -0.1036 | Yes | ||
| 132 | RRAD | 1422562_at | 21885 | -3.472 | -0.0693 | Yes | ||
| 133 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 21890 | -3.497 | -0.0345 | Yes | ||
| 134 | FOS | 1423100_at | 21901 | -3.644 | 0.0015 | Yes |

