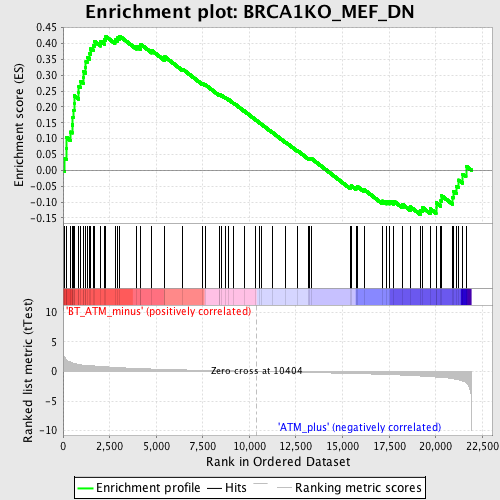
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | BRCA1KO_MEF_DN |
| Enrichment Score (ES) | 0.42343736 |
| Normalized Enrichment Score (NES) | 1.6725811 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2718 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 84 | 2.405 | 0.0386 | Yes | ||
| 2 | ROCK2 | 1423592_at 1451041_at | 175 | 1.980 | 0.0694 | Yes | ||
| 3 | GFRA1 | 1421973_at 1439015_at 1450440_at | 184 | 1.963 | 0.1037 | Yes | ||
| 4 | ANGEL2 | 1415731_at 1431606_a_at 1437050_s_at 1448360_s_at 1453741_x_at 1455362_at | 411 | 1.568 | 0.1210 | Yes | ||
| 5 | SMARCA3 | 1425996_a_at 1450889_at 1451901_at | 484 | 1.469 | 0.1437 | Yes | ||
| 6 | MGA | 1422646_at 1431232_a_at 1433012_at 1434746_at 1442356_at | 521 | 1.423 | 0.1671 | Yes | ||
| 7 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 562 | 1.372 | 0.1895 | Yes | ||
| 8 | USP47 | 1426976_at 1426977_at 1457454_at | 590 | 1.351 | 0.2121 | Yes | ||
| 9 | MAP2K4 | 1426233_at 1442679_at 1451982_at | 597 | 1.346 | 0.2356 | Yes | ||
| 10 | NCOR1 | 1423200_at 1423201_at 1423202_a_at 1435914_at 1438276_at 1444760_at 1445230_at 1446574_at 1457007_at 1457938_at 1458103_at | 840 | 1.172 | 0.2452 | Yes | ||
| 11 | UBE3A | 1416680_at 1416681_at 1416682_at 1425206_a_at 1431224_at 1445727_at | 846 | 1.170 | 0.2656 | Yes | ||
| 12 | EPN2 | 1427225_at 1427226_at 1433915_s_at 1440175_at 1447599_x_at 1454303_at 1454304_at | 941 | 1.124 | 0.2812 | Yes | ||
| 13 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 1077 | 1.067 | 0.2938 | Yes | ||
| 14 | UTX | 1427235_at 1427672_a_at 1440524_at 1441201_at 1445198_at 1446234_at 1446278_at | 1086 | 1.064 | 0.3123 | Yes | ||
| 15 | NFE2L2 | 1416543_at 1457117_at | 1211 | 1.028 | 0.3247 | Yes | ||
| 16 | CDC2L6 | 1434997_at 1442225_at | 1225 | 1.025 | 0.3422 | Yes | ||
| 17 | NCOA1 | 1418594_a_at 1434515_at 1439883_at 1443397_at 1443438_at 1459277_at | 1289 | 1.012 | 0.3572 | Yes | ||
| 18 | ZFR | 1419650_at 1442311_at 1443002_at 1443353_at 1443357_at 1444096_at 1449552_at 1458450_at | 1431 | 0.984 | 0.3681 | Yes | ||
| 19 | CHD1 | 1420990_at 1444787_at 1444788_x_at 1450077_at 1457473_at | 1480 | 0.974 | 0.3831 | Yes | ||
| 20 | COL3A1 | 1427883_a_at 1427884_at 1442977_at | 1614 | 0.945 | 0.3937 | Yes | ||
| 21 | ITCH | 1415769_at 1431316_at 1459332_at | 1688 | 0.928 | 0.4067 | Yes | ||
| 22 | NET1 | 1421321_a_at 1457830_at | 2008 | 0.853 | 0.4072 | Yes | ||
| 23 | SHOC2 | 1423129_at 1425845_a_at | 2217 | 0.805 | 0.4119 | Yes | ||
| 24 | VPS35 | 1415783_at 1415784_at | 2295 | 0.787 | 0.4223 | Yes | ||
| 25 | CLK1 | 1426124_a_at | 2787 | 0.684 | 0.4119 | Yes | ||
| 26 | YME1L1 | 1423360_at 1423361_at 1437822_at 1450954_at | 2914 | 0.663 | 0.4178 | Yes | ||
| 27 | PNN | 1423324_at 1423325_at | 3038 | 0.639 | 0.4234 | Yes | ||
| 28 | HOXB9 | 1452317_at | 3932 | 0.507 | 0.3915 | No | ||
| 29 | SEPP1 | 1452141_a_at | 4154 | 0.478 | 0.3899 | No | ||
| 30 | SLK | 1419092_a_at 1425977_a_at 1433999_at 1441262_at 1443815_x_at 1449336_a_at | 4157 | 0.478 | 0.3982 | No | ||
| 31 | ABCA1 | 1421839_at 1421840_at 1450392_at | 4772 | 0.413 | 0.3774 | No | ||
| 32 | EIF3S10 | 1416659_at 1416660_at 1416661_at 1448425_at | 5435 | 0.349 | 0.3533 | No | ||
| 33 | GOLGA4 | 1417674_s_at 1448803_at 1460213_at | 5463 | 0.347 | 0.3582 | No | ||
| 34 | CEBPZ | 1420497_a_at 1429962_at | 6436 | 0.266 | 0.3184 | No | ||
| 35 | ADD3 | 1423297_at 1423298_at 1426574_a_at 1447077_at 1456162_x_at | 7495 | 0.189 | 0.2733 | No | ||
| 36 | ZFX | 1419565_a_at 1425972_a_at 1428046_a_at 1449512_a_at | 7631 | 0.179 | 0.2703 | No | ||
| 37 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 8402 | 0.126 | 0.2373 | No | ||
| 38 | RABEP1 | 1421305_x_at 1426023_a_at 1427448_at 1443856_at 1444930_at 1459494_at | 8413 | 0.126 | 0.2391 | No | ||
| 39 | TGFBR3 | 1425620_at 1433795_at 1440041_at 1442332_at 1447314_at 1459355_at | 8523 | 0.119 | 0.2362 | No | ||
| 40 | HFE | 1422645_at 1450702_at | 8706 | 0.107 | 0.2298 | No | ||
| 41 | NKTR | 1423248_at 1423249_at 1437660_at 1440804_at 1458676_at | 8901 | 0.095 | 0.2226 | No | ||
| 42 | CASP1 | 1449265_at | 9164 | 0.079 | 0.2120 | No | ||
| 43 | CASP4 | 1449591_at | 9714 | 0.044 | 0.1877 | No | ||
| 44 | BAAT | 1419001_at 1419002_s_at | 10316 | 0.006 | 0.1603 | No | ||
| 45 | TJP1 | 1417749_a_at | 10543 | -0.008 | 0.1501 | No | ||
| 46 | KLF9 | 1422264_s_at 1428288_at 1428289_at 1436763_a_at 1436952_at 1452723_at 1456341_a_at | 10672 | -0.016 | 0.1445 | No | ||
| 47 | IPO7 | 1442511_at 1454955_at 1458165_at | 11255 | -0.054 | 0.1188 | No | ||
| 48 | GSPT1 | 1426736_at 1426737_at 1435000_at 1446550_at 1452168_x_at 1455173_at | 11964 | -0.097 | 0.0881 | No | ||
| 49 | TPP2 | 1421893_a_at 1421894_a_at 1430575_a_at 1430576_at 1438264_a_at | 12566 | -0.134 | 0.0630 | No | ||
| 50 | ZNF148 | 1418381_at 1427730_a_at 1436217_at 1441612_at 1441701_at 1449068_at 1449069_at 1455483_at | 13198 | -0.174 | 0.0372 | No | ||
| 51 | BNIP3 | 1422470_at | 13233 | -0.176 | 0.0388 | No | ||
| 52 | USP16 | 1430203_at 1431951_a_at | 13354 | -0.184 | 0.0365 | No | ||
| 53 | IMMT | 1429533_at 1429534_a_at 1433470_a_at 1437126_at | 15434 | -0.332 | -0.0527 | No | ||
| 54 | NID1 | 1416808_at 1441519_at | 15459 | -0.334 | -0.0479 | No | ||
| 55 | RAI14 | 1417400_at 1417401_at 1441030_at 1444777_at | 15729 | -0.353 | -0.0540 | No | ||
| 56 | LAMC1 | 1423885_at 1423886_at | 15810 | -0.360 | -0.0513 | No | ||
| 57 | MPO | 1415960_at | 16160 | -0.390 | -0.0604 | No | ||
| 58 | TLOC1 | 1432635_a_at 1433704_s_at 1439233_at 1444811_at 1454697_at 1455698_at | 17139 | -0.483 | -0.0966 | No | ||
| 59 | DCLRE1A | 1417904_at 1432097_a_at 1442329_at | 17349 | -0.505 | -0.0973 | No | ||
| 60 | NEFL | 1426255_at | 17545 | -0.526 | -0.0969 | No | ||
| 61 | VLDLR | 1417900_a_at 1434465_x_at 1435893_at 1438258_at 1442169_at 1451156_s_at | 17758 | -0.551 | -0.0969 | No | ||
| 62 | FAF1 | 1418242_at 1434370_s_at | 18213 | -0.611 | -0.1068 | No | ||
| 63 | UBE2L3 | 1417907_at 1417908_s_at 1448879_at 1448880_at | 18651 | -0.675 | -0.1149 | No | ||
| 64 | GSTA4 | 1416368_at | 19183 | -0.761 | -0.1258 | No | ||
| 65 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 19284 | -0.782 | -0.1166 | No | ||
| 66 | GPT2 | 1434542_at 1438385_s_at 1439029_at 1455007_s_at | 19721 | -0.867 | -0.1212 | No | ||
| 67 | PDIA4 | 1416497_at | 20027 | -0.938 | -0.1186 | No | ||
| 68 | SMAD1 | 1416081_at 1448208_at 1459843_s_at | 20029 | -0.939 | -0.1021 | No | ||
| 69 | BIRC3 | 1421392_a_at 1425223_at | 20286 | -1.002 | -0.0961 | No | ||
| 70 | ENPP2 | 1415894_at 1443224_at 1448136_at | 20328 | -1.009 | -0.0802 | No | ||
| 71 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20910 | -1.212 | -0.0854 | No | ||
| 72 | XRN2 | 1422842_at 1450777_at | 20952 | -1.232 | -0.0655 | No | ||
| 73 | NDRG3 | 1417663_a_at 1417664_a_at | 21135 | -1.326 | -0.0504 | No | ||
| 74 | CCPG1 | 1420127_s_at 1424420_at 1447003_at | 21243 | -1.410 | -0.0304 | No | ||
| 75 | STK10 | 1417751_at 1459736_at | 21460 | -1.615 | -0.0118 | No | ||
| 76 | BAZ2B | 1435240_at 1440984_at 1441556_at 1442704_at 1442971_at 1445048_at 1448007_at 1458077_at | 21636 | -1.895 | 0.0136 | No |

