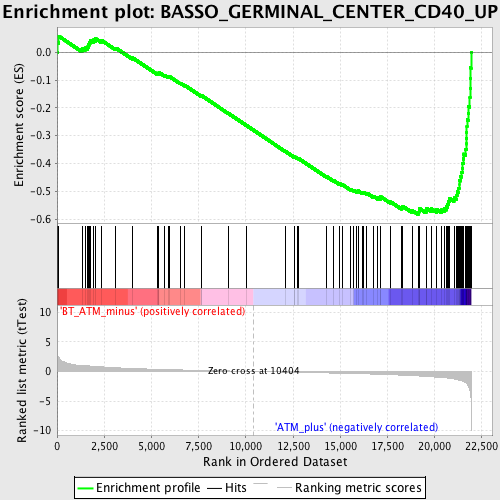
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | BASSO_GERMINAL_CENTER_CD40_UP |
| Enrichment Score (ES) | -0.58361787 |
| Normalized Enrichment Score (NES) | -2.209722 |
| Nominal p-value | 0.0 |
| FDR q-value | 9.25613E-4 |
| FWER p-Value | 0.0050 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2B | 1434653_at 1442437_at 1442927_at | 16 | 3.364 | 0.0355 | No | ||
| 2 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 84 | 2.405 | 0.0584 | No | ||
| 3 | TAP1 | 1416016_at 1420274_at 1448177_at 1449803_x_at | 1320 | 1.006 | 0.0127 | No | ||
| 4 | CHD1 | 1420990_at 1444787_at 1444788_x_at 1450077_at 1457473_at | 1480 | 0.974 | 0.0159 | No | ||
| 5 | PLEK | 1417523_at 1448748_at 1448749_at | 1628 | 0.942 | 0.0194 | No | ||
| 6 | LAPTM5 | 1426025_s_at 1436905_x_at 1447742_at 1459841_x_at | 1677 | 0.930 | 0.0272 | No | ||
| 7 | HSP90B1 | 1415889_a_at 1438040_a_at | 1697 | 0.926 | 0.0363 | No | ||
| 8 | IER3 | 1419647_a_at | 1760 | 0.910 | 0.0433 | No | ||
| 9 | SLAMF1 | 1425569_a_at 1425570_at 1425571_at | 1937 | 0.871 | 0.0446 | No | ||
| 10 | SH2B3 | 1433455_at 1442756_at 1450183_a_at | 2042 | 0.846 | 0.0490 | No | ||
| 11 | SNF1LK | 1419766_at 1459601_at | 2335 | 0.779 | 0.0440 | No | ||
| 12 | NFKBIE | 1431843_a_at 1458299_s_at | 3109 | 0.625 | 0.0154 | No | ||
| 13 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 3986 | 0.501 | -0.0193 | No | ||
| 14 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 5294 | 0.363 | -0.0752 | No | ||
| 15 | CCL4 | 1421578_at | 5373 | 0.355 | -0.0750 | No | ||
| 16 | PTP4A3 | 1418181_at 1418182_at | 5393 | 0.354 | -0.0720 | No | ||
| 17 | LGALS1 | 1419573_a_at 1455439_a_at | 5692 | 0.327 | -0.0821 | No | ||
| 18 | TCFL5 | 1456515_s_at | 5876 | 0.310 | -0.0872 | No | ||
| 19 | CR2 | 1425289_a_at | 5962 | 0.303 | -0.0878 | No | ||
| 20 | RGS1 | 1417601_at | 6520 | 0.259 | -0.1105 | No | ||
| 21 | HTR3A | 1418268_at | 6757 | 0.242 | -0.1187 | No | ||
| 22 | RAB27A | 1425284_a_at 1425285_a_at 1429123_at 1454438_at | 7668 | 0.176 | -0.1584 | No | ||
| 23 | CCL3 | 1419561_at | 7670 | 0.176 | -0.1566 | No | ||
| 24 | NEF3 | 1422520_at | 9058 | 0.086 | -0.2191 | No | ||
| 25 | RXRA | 1425762_a_at 1430497_at 1454773_at | 10011 | 0.025 | -0.2624 | No | ||
| 26 | LHFPL2 | 1434129_s_at 1434130_at 1443185_at 1454847_at | 12078 | -0.103 | -0.3559 | No | ||
| 27 | TRIP10 | 1418092_s_at | 12569 | -0.134 | -0.3769 | No | ||
| 28 | PRG1 | 1417426_at | 12592 | -0.135 | -0.3764 | No | ||
| 29 | MAP3K8 | 1419208_at | 12729 | -0.144 | -0.3811 | No | ||
| 30 | NCF2 | 1448561_at | 12797 | -0.149 | -0.3825 | No | ||
| 31 | TNFAIP8 | 1416950_at 1442239_at 1442753_at 1444224_at 1457996_at | 14284 | -0.247 | -0.4479 | No | ||
| 32 | TNFAIP2 | 1416273_at 1438855_x_at 1457918_at | 14627 | -0.273 | -0.4606 | No | ||
| 33 | CFLAR | 1424996_at 1425686_at 1425687_at 1439600_at 1449317_at | 14961 | -0.297 | -0.4726 | No | ||
| 34 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 15109 | -0.307 | -0.4761 | No | ||
| 35 | IL1B | 1449399_a_at | 15545 | -0.340 | -0.4923 | No | ||
| 36 | PHACTR1 | 1432851_at 1432852_at 1439022_at 1454832_at | 15699 | -0.352 | -0.4955 | No | ||
| 37 | ARID5B | 1420972_at 1420973_at 1434283_at 1442176_at 1445423_at 1456831_at 1456973_at 1457483_at 1458238_at | 15869 | -0.366 | -0.4993 | No | ||
| 38 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 15936 | -0.372 | -0.4983 | No | ||
| 39 | MAPKAPK2 | 1426648_at | 16184 | -0.392 | -0.5054 | No | ||
| 40 | DTX2 | 1421720_a_at 1439429_x_at | 16236 | -0.397 | -0.5034 | No | ||
| 41 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 16399 | -0.411 | -0.5064 | No | ||
| 42 | NFKB2 | 1425902_a_at 1429128_x_at | 16780 | -0.446 | -0.5190 | No | ||
| 43 | STAT5A | 1421469_a_at 1450259_a_at | 16987 | -0.467 | -0.5234 | No | ||
| 44 | SYNGR2 | 1417081_a_at 1448577_x_at | 17115 | -0.480 | -0.5240 | No | ||
| 45 | SNN | 1422310_at 1452789_at | 17141 | -0.483 | -0.5200 | No | ||
| 46 | ELL2 | 1450744_at 1458569_at | 17670 | -0.541 | -0.5383 | No | ||
| 47 | BLR1 | 1422003_at | 18236 | -0.615 | -0.5575 | No | ||
| 48 | RAC2 | 1417620_at 1440208_at | 18302 | -0.623 | -0.5538 | No | ||
| 49 | ICOSLG | 1419212_at 1426087_at | 18803 | -0.698 | -0.5692 | No | ||
| 50 | TNF | 1419607_at | 19120 | -0.750 | -0.5755 | Yes | ||
| 51 | PLXNC1 | 1423213_at 1423214_at 1443433_at 1443434_s_at 1450905_at 1450906_at | 19166 | -0.757 | -0.5694 | Yes | ||
| 52 | TMSB4X | 1415906_at | 19207 | -0.768 | -0.5630 | Yes | ||
| 53 | EHD1 | 1416010_a_at 1416011_x_at 1416012_at 1448175_at | 19541 | -0.829 | -0.5693 | Yes | ||
| 54 | KCNN4 | 1421038_a_at 1435945_a_at | 19562 | -0.833 | -0.5612 | Yes | ||
| 55 | IRF5 | 1460231_at | 19814 | -0.887 | -0.5631 | Yes | ||
| 56 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 20105 | -0.957 | -0.5661 | Yes | ||
| 57 | TNFAIP3 | 1433699_at 1450829_at | 20369 | -1.017 | -0.5672 | Yes | ||
| 58 | ADAM8 | 1416871_at | 20521 | -1.054 | -0.5627 | Yes | ||
| 59 | LITAF | 1416303_at 1416304_at | 20628 | -1.087 | -0.5558 | Yes | ||
| 60 | TNFSF9 | 1422924_at | 20676 | -1.101 | -0.5461 | Yes | ||
| 61 | UNC119 | 1418123_at | 20725 | -1.123 | -0.5362 | Yes | ||
| 62 | NCKAP1L | 1428786_at 1428787_at 1458516_at | 20779 | -1.146 | -0.5263 | Yes | ||
| 63 | CAPG | 1447803_x_at 1450355_a_at | 21021 | -1.265 | -0.5237 | Yes | ||
| 64 | DDIT4 | 1428306_at | 21169 | -1.350 | -0.5158 | Yes | ||
| 65 | FAIM3 | 1429889_at | 21186 | -1.362 | -0.5019 | Yes | ||
| 66 | ZFP36L1 | 1422528_a_at 1450644_at | 21264 | -1.433 | -0.4900 | Yes | ||
| 67 | MTMR4 | 1418150_at 1418151_at 1459874_s_at | 21289 | -1.452 | -0.4754 | Yes | ||
| 68 | TCEB3 | 1422591_at 1434117_at 1450676_at | 21292 | -1.453 | -0.4598 | Yes | ||
| 69 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 21365 | -1.518 | -0.4468 | Yes | ||
| 70 | ICAM1 | 1424067_at | 21412 | -1.572 | -0.4319 | Yes | ||
| 71 | STK10 | 1417751_at 1459736_at | 21460 | -1.615 | -0.4167 | Yes | ||
| 72 | TMC6 | 1423770_at | 21482 | -1.642 | -0.3999 | Yes | ||
| 73 | IRF4 | 1421173_at 1421174_at | 21520 | -1.685 | -0.3835 | Yes | ||
| 74 | LTA | 1420353_at | 21540 | -1.710 | -0.3659 | Yes | ||
| 75 | BATF | 1419410_at | 21644 | -1.915 | -0.3500 | Yes | ||
| 76 | CD40 | 1439221_s_at 1449473_s_at 1460415_a_at | 21666 | -1.970 | -0.3297 | Yes | ||
| 77 | JUNB | 1415899_at | 21672 | -1.992 | -0.3084 | Yes | ||
| 78 | DUSP2 | 1450698_at | 21696 | -2.054 | -0.2873 | Yes | ||
| 79 | TRAF1 | 1423602_at 1445452_at | 21704 | -2.081 | -0.2652 | Yes | ||
| 80 | CD74 | 1425519_a_at | 21711 | -2.114 | -0.2427 | Yes | ||
| 81 | IFI30 | 1422476_at | 21770 | -2.329 | -0.2202 | Yes | ||
| 82 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21794 | -2.532 | -0.1940 | Yes | ||
| 83 | CD83 | 1416111_at | 21863 | -3.155 | -0.1631 | Yes | ||
| 84 | CCL22 | 1417925_at | 21871 | -3.238 | -0.1285 | Yes | ||
| 85 | EBI2 | 1437356_at 1457691_at | 21881 | -3.381 | -0.0925 | Yes | ||
| 86 | ISGF3G | 1421322_a_at | 21889 | -3.496 | -0.0551 | Yes | ||
| 87 | CCR7 | 1423466_at | 21924 | -5.296 | 0.0005 | Yes |

