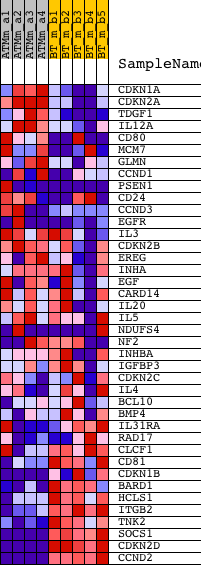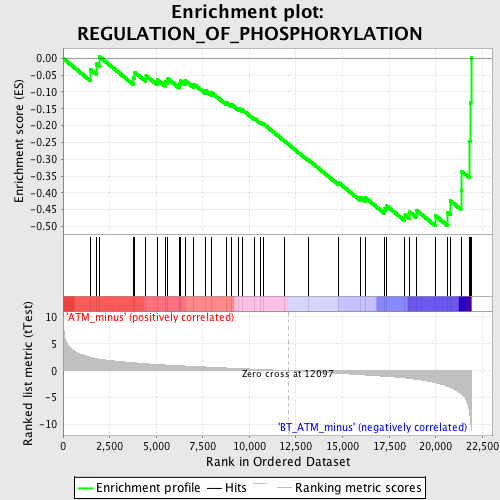
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | REGULATION_OF_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.49869642 |
| Normalized Enrichment Score (NES) | -1.631399 |
| Nominal p-value | 0.00754717 |
| FDR q-value | 0.47120655 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN1A | 1421679_a_at 1424638_at | 1464 | 2.524 | -0.0327 | No | ||
| 2 | CDKN2A | 1450140_a_at | 1778 | 2.279 | -0.0161 | No | ||
| 3 | TDGF1 | 1450989_at | 1963 | 2.151 | 0.0046 | No | ||
| 4 | IL12A | 1425454_a_at | 3763 | 1.492 | -0.0574 | No | ||
| 5 | CD80 | 1427717_at 1432826_a_at 1440592_at 1451950_a_at 1454372_at 1457952_at | 3845 | 1.466 | -0.0413 | No | ||
| 6 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 4450 | 1.300 | -0.0512 | No | ||
| 7 | GLMN | 1418839_at | 5047 | 1.153 | -0.0629 | No | ||
| 8 | CCND1 | 1417419_at 1417420_at 1448698_at | 5473 | 1.073 | -0.0677 | No | ||
| 9 | PSEN1 | 1420261_at 1421853_at 1425549_at 1450399_at | 5624 | 1.041 | -0.0605 | No | ||
| 10 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 6232 | 0.937 | -0.0756 | No | ||
| 11 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 6317 | 0.920 | -0.0669 | No | ||
| 12 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 6570 | 0.873 | -0.0666 | No | ||
| 13 | IL3 | 1450566_at | 7020 | 0.793 | -0.0764 | No | ||
| 14 | CDKN2B | 1449152_at | 7628 | 0.694 | -0.0947 | No | ||
| 15 | EREG | 1419431_at | 7953 | 0.640 | -0.1009 | No | ||
| 16 | INHA | 1422728_at | 8763 | 0.512 | -0.1309 | No | ||
| 17 | EGF | 1418093_a_at | 9052 | 0.465 | -0.1377 | No | ||
| 18 | CARD14 | 1450142_a_at | 9437 | 0.408 | -0.1498 | No | ||
| 19 | IL20 | 1421608_at | 9612 | 0.384 | -0.1525 | No | ||
| 20 | IL5 | 1425546_a_at 1442049_at 1450550_at | 10259 | 0.282 | -0.1782 | No | ||
| 21 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 10577 | 0.234 | -0.1895 | No | ||
| 22 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 10753 | 0.207 | -0.1947 | No | ||
| 23 | INHBA | 1422053_at 1458291_at | 11897 | 0.031 | -0.2465 | No | ||
| 24 | IGFBP3 | 1423062_at 1458268_s_at | 13172 | -0.184 | -0.3022 | No | ||
| 25 | CDKN2C | 1416868_at 1439164_at | 14766 | -0.472 | -0.3685 | No | ||
| 26 | IL4 | 1449864_at | 15987 | -0.733 | -0.4143 | No | ||
| 27 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 16226 | -0.792 | -0.4145 | No | ||
| 28 | BMP4 | 1422912_at | 17235 | -1.026 | -0.4466 | No | ||
| 29 | IL31RA | 1445095_at 1451535_at | 17340 | -1.045 | -0.4372 | No | ||
| 30 | RAD17 | 1448762_at | 18358 | -1.353 | -0.4654 | No | ||
| 31 | CLCF1 | 1437270_a_at 1437271_at 1450262_at | 18607 | -1.452 | -0.4570 | No | ||
| 32 | CD81 | 1416330_at | 18963 | -1.608 | -0.4515 | No | ||
| 33 | CDKN1B | 1419497_at 1434045_at | 19998 | -2.247 | -0.4683 | Yes | ||
| 34 | BARD1 | 1420594_at | 20634 | -2.871 | -0.4584 | Yes | ||
| 35 | HCLS1 | 1418842_at | 20791 | -3.090 | -0.4237 | Yes | ||
| 36 | ITGB2 | 1450678_at | 21367 | -4.241 | -0.3926 | Yes | ||
| 37 | TNK2 | 1448298_at | 21388 | -4.289 | -0.3354 | Yes | ||
| 38 | SOCS1 | 1440047_at 1450446_a_at | 21837 | -8.094 | -0.2463 | Yes | ||
| 39 | CDKN2D | 1416253_at | 21858 | -8.553 | -0.1315 | Yes | ||
| 40 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21904 | -9.965 | 0.0014 | Yes |