Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
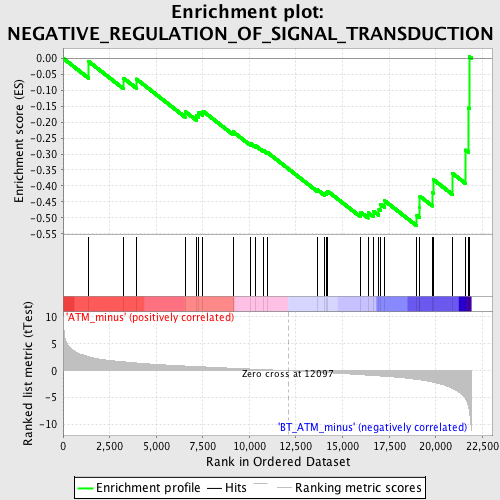
| GeneSet | NEGATIVE_REGULATION_OF_SIGNAL_TRANSDUCTION |
| Enrichment Score (ES) | -0.5254447 |
| Normalized Enrichment Score (NES) | -1.6501052 |
| Nominal p-value | 0.013232514 |
| FDR q-value | 0.54387224 |
| FWER p-Value | 1.0 |

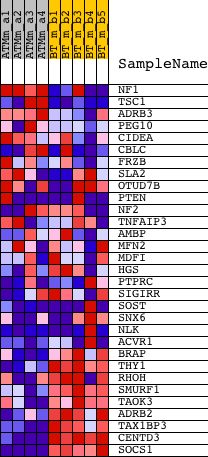
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 1365 | 2.618 | -0.0098 | No | ||
| 2 | TSC1 | 1422043_at 1439989_at 1455252_at | 3251 | 1.648 | -0.0628 | No | ||
| 3 | ADRB3 | 1421555_at 1455918_at | 3934 | 1.444 | -0.0650 | No | ||
| 4 | PEG10 | 1427550_at | 6551 | 0.876 | -0.1668 | No | ||
| 5 | CIDEA | 1417956_at | 7154 | 0.771 | -0.1788 | No | ||
| 6 | CBLC | 1422666_at 1431692_a_at | 7290 | 0.749 | -0.1700 | No | ||
| 7 | FRZB | 1416658_at 1448424_at | 7509 | 0.714 | -0.1656 | No | ||
| 8 | SLA2 | 1437504_at 1451492_at | 9126 | 0.454 | -0.2303 | No | ||
| 9 | OTUD7B | 1428953_at 1429139_at 1435552_at 1457716_at | 10063 | 0.311 | -0.2668 | No | ||
| 10 | PTEN | 1422553_at 1441593_at 1444325_at 1450655_at 1454722_at 1455728_at 1457493_at | 10343 | 0.267 | -0.2742 | No | ||
| 11 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 10753 | 0.207 | -0.2887 | No | ||
| 12 | TNFAIP3 | 1433699_at 1450829_at | 10976 | 0.174 | -0.2953 | No | ||
| 13 | AMBP | 1416649_at | 13647 | -0.268 | -0.4118 | No | ||
| 14 | MFN2 | 1441424_at 1448131_at 1451900_at | 14037 | -0.333 | -0.4229 | No | ||
| 15 | MDFI | 1420713_a_at 1425924_at | 14130 | -0.348 | -0.4201 | No | ||
| 16 | HGS | 1416540_at | 14208 | -0.363 | -0.4164 | No | ||
| 17 | PTPRC | 1422124_a_at 1440165_at | 15964 | -0.730 | -0.4819 | No | ||
| 18 | SIGIRR | 1449163_at | 16374 | -0.830 | -0.4839 | No | ||
| 19 | SOST | 1421245_at 1436240_at 1450179_at | 16666 | -0.904 | -0.4790 | No | ||
| 20 | SNX6 | 1425148_a_at 1429497_s_at 1447234_s_at 1451602_at | 16960 | -0.968 | -0.4730 | No | ||
| 21 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 17047 | -0.988 | -0.4571 | No | ||
| 22 | ACVR1 | 1416786_at 1416787_at 1448460_at 1457551_at | 17261 | -1.031 | -0.4461 | No | ||
| 23 | BRAP | 1416738_at 1416739_a_at | 18999 | -1.623 | -0.4929 | Yes | ||
| 24 | THY1 | 1423135_at | 19157 | -1.688 | -0.4662 | Yes | ||
| 25 | RHOH | 1429319_at 1441385_at 1443015_at | 19162 | -1.691 | -0.4324 | Yes | ||
| 26 | SMURF1 | 1443393_at | 19832 | -2.107 | -0.4207 | Yes | ||
| 27 | TAOK3 | 1435964_a_at 1444366_at 1444881_at 1447257_at 1455733_at | 19879 | -2.140 | -0.3799 | Yes | ||
| 28 | ADRB2 | 1437302_at | 20916 | -3.306 | -0.3608 | Yes | ||
| 29 | TAX1BP3 | 1424169_at | 21617 | -5.254 | -0.2874 | Yes | ||
| 30 | CENTD3 | 1419833_s_at 1447407_at 1451282_at | 21787 | -6.947 | -0.1557 | Yes | ||
| 31 | SOCS1 | 1440047_at 1450446_a_at | 21837 | -8.094 | 0.0044 | Yes |