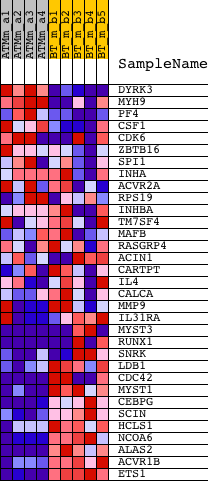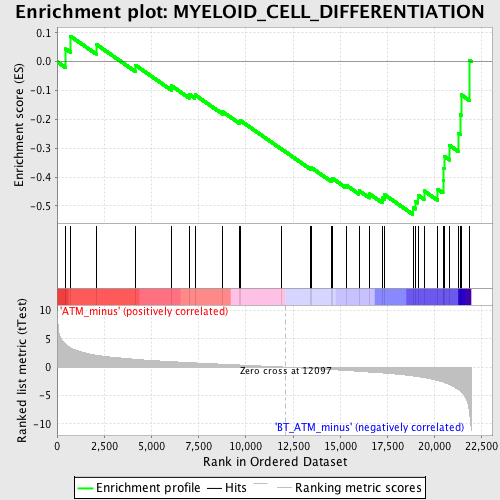
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | MYELOID_CELL_DIFFERENTIATION |
| Enrichment Score (ES) | -0.5297627 |
| Normalized Enrichment Score (NES) | -1.6755266 |
| Nominal p-value | 0.0056179776 |
| FDR q-value | 0.60590523 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DYRK3 | 1424229_at | 458 | 4.002 | 0.0442 | No | ||
| 2 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 702 | 3.417 | 0.0887 | No | ||
| 3 | PF4 | 1448995_at | 2066 | 2.093 | 0.0606 | No | ||
| 4 | CSF1 | 1425154_a_at 1425155_x_at 1448914_a_at 1460220_a_at | 4141 | 1.383 | -0.0116 | No | ||
| 5 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 6050 | 0.964 | -0.0830 | No | ||
| 6 | ZBTB16 | 1419874_x_at 1427638_at 1439163_at 1445562_at 1456986_at 1459557_at | 6994 | 0.798 | -0.1131 | No | ||
| 7 | SPI1 | 1418747_at | 7302 | 0.747 | -0.1150 | No | ||
| 8 | INHA | 1422728_at | 8763 | 0.512 | -0.1733 | No | ||
| 9 | ACVR2A | 1437382_at 1439643_at 1445026_at 1451004_at | 9664 | 0.377 | -0.2083 | No | ||
| 10 | RPS19 | 1449243_a_at 1460442_at | 9687 | 0.373 | -0.2032 | No | ||
| 11 | INHBA | 1422053_at 1458291_at | 11897 | 0.031 | -0.3035 | No | ||
| 12 | TM7SF4 | 1431970_at | 13409 | -0.224 | -0.3689 | No | ||
| 13 | MAFB | 1451715_at 1451716_at | 13457 | -0.233 | -0.3672 | No | ||
| 14 | RASGRP4 | 1425380_at | 14520 | -0.423 | -0.4088 | No | ||
| 15 | ACIN1 | 1416568_a_at 1421197_a_at | 14572 | -0.433 | -0.4041 | No | ||
| 16 | CARTPT | 1422825_at | 15309 | -0.585 | -0.4282 | No | ||
| 17 | IL4 | 1449864_at | 15987 | -0.733 | -0.4472 | No | ||
| 18 | CALCA | 1427355_at 1452004_at | 16545 | -0.871 | -0.4585 | No | ||
| 19 | MMP9 | 1416298_at 1448291_at | 17217 | -1.023 | -0.4724 | No | ||
| 20 | IL31RA | 1445095_at 1451535_at | 17340 | -1.045 | -0.4610 | No | ||
| 21 | MYST3 | 1436315_at 1437487_at 1446143_at 1446405_at | 18847 | -1.558 | -0.5044 | Yes | ||
| 22 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 18990 | -1.619 | -0.4845 | Yes | ||
| 23 | SNRK | 1425678_a_at 1448864_at | 19112 | -1.668 | -0.4629 | Yes | ||
| 24 | LDB1 | 1452024_a_at | 19436 | -1.817 | -0.4481 | Yes | ||
| 25 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 20162 | -2.391 | -0.4423 | Yes | ||
| 26 | MYST1 | 1432080_s_at | 20438 | -2.621 | -0.4122 | Yes | ||
| 27 | CEBPG | 1425261_at 1425262_at 1451639_at | 20471 | -2.660 | -0.3703 | Yes | ||
| 28 | SCIN | 1450276_a_at | 20499 | -2.694 | -0.3277 | Yes | ||
| 29 | HCLS1 | 1418842_at | 20791 | -3.090 | -0.2907 | Yes | ||
| 30 | NCOA6 | 1423374_at | 21241 | -3.923 | -0.2473 | Yes | ||
| 31 | ALAS2 | 1451675_a_at | 21365 | -4.230 | -0.1841 | Yes | ||
| 32 | ACVR1B | 1422098_at 1433725_at | 21411 | -4.378 | -0.1149 | Yes | ||
| 33 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 21857 | -8.522 | 0.0035 | Yes |