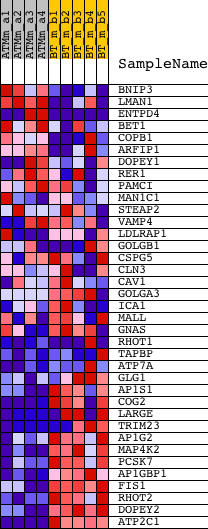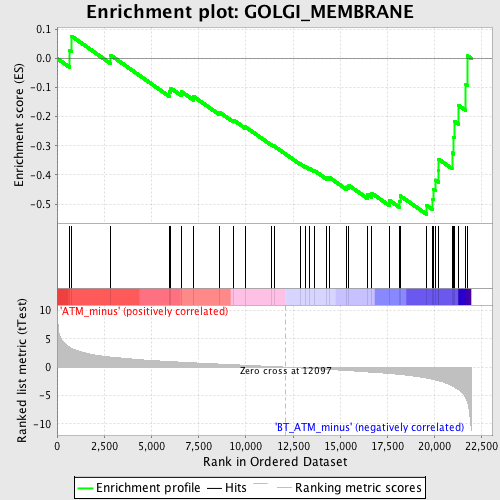
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GOLGI_MEMBRANE |
| Enrichment Score (ES) | -0.5358722 |
| Normalized Enrichment Score (NES) | -1.7481074 |
| Nominal p-value | 0.0019120459 |
| FDR q-value | 0.7328477 |
| FWER p-Value | 0.937 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BNIP3 | 1422470_at | 649 | 3.529 | 0.0279 | No | ||
| 2 | LMAN1 | 1428129_at 1428130_at 1444037_at 1452671_s_at | 763 | 3.320 | 0.0769 | No | ||
| 3 | ENTPD4 | 1431761_at 1438177_x_at 1447900_x_at 1449190_a_at | 2822 | 1.779 | 0.0119 | No | ||
| 4 | BET1 | 1416866_at 1416867_at | 5940 | 0.984 | -0.1144 | No | ||
| 5 | COPB1 | 1423301_at | 6018 | 0.969 | -0.1021 | No | ||
| 6 | ARFIP1 | 1454916_s_at | 6595 | 0.869 | -0.1142 | No | ||
| 7 | DOPEY1 | 1435295_at 1442855_at | 7230 | 0.757 | -0.1308 | No | ||
| 8 | RER1 | 1460660_x_at 1460706_s_at | 8585 | 0.540 | -0.1838 | No | ||
| 9 | PAMCI | 1427942_at | 9338 | 0.423 | -0.2113 | No | ||
| 10 | MAN1C1 | 1436193_at | 9963 | 0.327 | -0.2344 | No | ||
| 11 | STEAP2 | 1428636_at 1438773_at 1442359_at 1444290_at 1446071_at | 11352 | 0.115 | -0.2959 | No | ||
| 12 | VAMP4 | 1422895_at 1422896_at 1434796_at | 11486 | 0.096 | -0.3004 | No | ||
| 13 | LDLRAP1 | 1424378_at 1457162_at | 12866 | -0.133 | -0.3613 | No | ||
| 14 | GOLGB1 | 1433135_at 1435032_at 1460535_at | 13141 | -0.179 | -0.3708 | No | ||
| 15 | CSPG5 | 1450713_at | 13345 | -0.214 | -0.3766 | No | ||
| 16 | CLN3 | 1417551_at 1457789_at | 13611 | -0.260 | -0.3845 | No | ||
| 17 | CAV1 | 1449145_a_at | 14284 | -0.376 | -0.4090 | No | ||
| 18 | GOLGA3 | 1419159_at 1419160_at 1449616_s_at | 14405 | -0.400 | -0.4080 | No | ||
| 19 | ICA1 | 1417901_a_at 1431644_a_at | 15350 | -0.593 | -0.4414 | No | ||
| 20 | MALL | 1425359_at 1435693_at | 15455 | -0.614 | -0.4362 | No | ||
| 21 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 16450 | -0.849 | -0.4677 | No | ||
| 22 | RHOT1 | 1423533_a_at 1440853_at | 16655 | -0.899 | -0.4623 | No | ||
| 23 | TAPBP | 1421812_at 1450378_at | 17595 | -1.113 | -0.4871 | No | ||
| 24 | ATP7A | 1418774_a_at 1436921_at | 18119 | -1.271 | -0.4902 | No | ||
| 25 | GLG1 | 1417087_at 1448579_at 1448580_at 1460554_s_at | 18166 | -1.286 | -0.4713 | No | ||
| 26 | AP1S1 | 1416087_at | 19580 | -1.906 | -0.5048 | Yes | ||
| 27 | COG2 | 1416932_at | 19898 | -2.164 | -0.4840 | Yes | ||
| 28 | LARGE | 1417435_at 1417436_at 1441642_at 1442420_at 1443212_at 1458663_at | 19917 | -2.184 | -0.4492 | Yes | ||
| 29 | TRIM23 | 1426969_at 1429998_at 1456089_at | 20038 | -2.287 | -0.4174 | Yes | ||
| 30 | AP1G2 | 1419113_at | 20203 | -2.420 | -0.3854 | Yes | ||
| 31 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 20211 | -2.423 | -0.3462 | Yes | ||
| 32 | PCSK7 | 1417180_at | 20917 | -3.310 | -0.3244 | Yes | ||
| 33 | AP1GBP1 | 1435215_at 1442344_at 1457695_at | 21006 | -3.474 | -0.2718 | Yes | ||
| 34 | FIS1 | 1416764_at 1443819_x_at | 21029 | -3.513 | -0.2155 | Yes | ||
| 35 | RHOT2 | 1426822_at | 21256 | -3.972 | -0.1610 | Yes | ||
| 36 | DOPEY2 | 1428330_at 1449904_at | 21648 | -5.506 | -0.0891 | Yes | ||
| 37 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 21738 | -6.260 | 0.0090 | Yes |