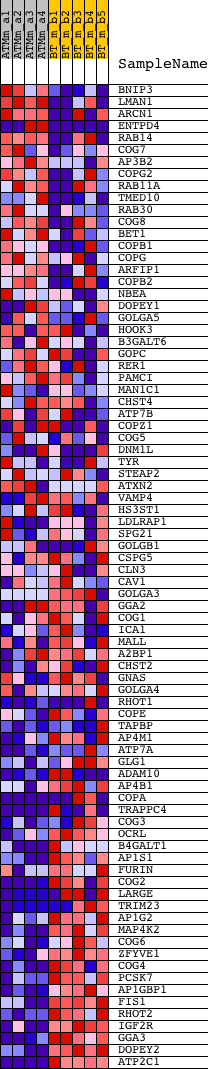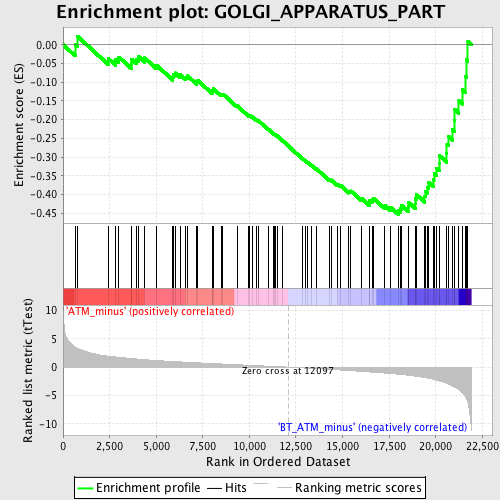
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GOLGI_APPARATUS_PART |
| Enrichment Score (ES) | -0.4535917 |
| Normalized Enrichment Score (NES) | -1.679301 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.6467931 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BNIP3 | 1422470_at | 649 | 3.529 | 0.0002 | No | ||
| 2 | LMAN1 | 1428129_at 1428130_at 1444037_at 1452671_s_at | 763 | 3.320 | 0.0231 | No | ||
| 3 | ARCN1 | 1423743_at 1436062_at 1444372_at 1451089_a_at | 2438 | 1.924 | -0.0372 | No | ||
| 4 | ENTPD4 | 1431761_at 1438177_x_at 1447900_x_at 1449190_a_at | 2822 | 1.779 | -0.0397 | No | ||
| 5 | RAB14 | 1415686_at 1419243_at 1419244_a_at 1419245_at 1419246_s_at 1439687_at 1441992_at | 2995 | 1.722 | -0.0330 | No | ||
| 6 | COG7 | 1443486_at | 3669 | 1.515 | -0.0510 | No | ||
| 7 | AP3B2 | 1426927_at 1445224_at 1451837_at | 3680 | 1.512 | -0.0387 | No | ||
| 8 | COPG2 | 1448761_a_at | 3958 | 1.437 | -0.0392 | No | ||
| 9 | RAB11A | 1449256_a_at | 4062 | 1.405 | -0.0320 | No | ||
| 10 | TMED10 | 1424707_at 1424708_at | 4378 | 1.320 | -0.0352 | No | ||
| 11 | RAB30 | 1426452_a_at | 5013 | 1.158 | -0.0545 | No | ||
| 12 | COG8 | 1420265_x_at 1420266_at 1423617_at 1426821_at 1447860_x_at 1451052_at | 5898 | 0.992 | -0.0865 | No | ||
| 13 | BET1 | 1416866_at 1416867_at | 5940 | 0.984 | -0.0801 | No | ||
| 14 | COPB1 | 1423301_at | 6018 | 0.969 | -0.0754 | No | ||
| 15 | COPG | 1415670_at 1416017_at | 6296 | 0.925 | -0.0802 | No | ||
| 16 | ARFIP1 | 1454916_s_at | 6595 | 0.869 | -0.0865 | No | ||
| 17 | COPB2 | 1452102_at 1456175_a_at | 6673 | 0.854 | -0.0828 | No | ||
| 18 | NBEA | 1452251_at | 7166 | 0.769 | -0.0988 | No | ||
| 19 | DOPEY1 | 1435295_at 1442855_at | 7230 | 0.757 | -0.0953 | No | ||
| 20 | GOLGA5 | 1418447_at | 8004 | 0.631 | -0.1253 | No | ||
| 21 | HOOK3 | 1443857_at 1446737_a_at | 8028 | 0.627 | -0.1211 | No | ||
| 22 | B3GALT6 | 1421155_at 1435252_at 1440340_at | 8075 | 0.618 | -0.1179 | No | ||
| 23 | GOPC | 1421191_s_at 1450153_at 1453221_at | 8480 | 0.558 | -0.1317 | No | ||
| 24 | RER1 | 1460660_x_at 1460706_s_at | 8585 | 0.540 | -0.1319 | No | ||
| 25 | PAMCI | 1427942_at | 9338 | 0.423 | -0.1627 | No | ||
| 26 | MAN1C1 | 1436193_at | 9963 | 0.327 | -0.1885 | No | ||
| 27 | CHST4 | 1453393_a_at | 10021 | 0.318 | -0.1884 | No | ||
| 28 | ATP7B | 1421563_at 1446004_at 1450283_at | 10162 | 0.298 | -0.1923 | No | ||
| 29 | COPZ1 | 1447834_at 1451825_a_at | 10358 | 0.265 | -0.1990 | No | ||
| 30 | COG5 | 1435298_at 1445806_at | 10486 | 0.246 | -0.2027 | No | ||
| 31 | DNM1L | 1428008_at 1428086_at 1428087_at 1452638_s_at | 11032 | 0.165 | -0.2263 | No | ||
| 32 | TYR | 1417717_a_at 1448821_at 1456095_at | 11309 | 0.121 | -0.2379 | No | ||
| 33 | STEAP2 | 1428636_at 1438773_at 1442359_at 1444290_at 1446071_at | 11352 | 0.115 | -0.2388 | No | ||
| 34 | ATXN2 | 1419866_s_at 1438143_s_at 1438144_x_at 1443516_at 1459363_at 1460653_at | 11424 | 0.105 | -0.2412 | No | ||
| 35 | VAMP4 | 1422895_at 1422896_at 1434796_at | 11486 | 0.096 | -0.2432 | No | ||
| 36 | HS3ST1 | 1423450_a_at | 11786 | 0.048 | -0.2564 | No | ||
| 37 | LDLRAP1 | 1424378_at 1457162_at | 12866 | -0.133 | -0.3047 | No | ||
| 38 | SPG21 | 1451036_at | 13036 | -0.164 | -0.3110 | No | ||
| 39 | GOLGB1 | 1433135_at 1435032_at 1460535_at | 13141 | -0.179 | -0.3143 | No | ||
| 40 | CSPG5 | 1450713_at | 13345 | -0.214 | -0.3218 | No | ||
| 41 | CLN3 | 1417551_at 1457789_at | 13611 | -0.260 | -0.3317 | No | ||
| 42 | CAV1 | 1449145_a_at | 14284 | -0.376 | -0.3593 | No | ||
| 43 | GOLGA3 | 1419159_at 1419160_at 1449616_s_at | 14405 | -0.400 | -0.3614 | No | ||
| 44 | GGA2 | 1428141_at 1457955_at | 14757 | -0.470 | -0.3734 | No | ||
| 45 | COG1 | 1418354_at 1433774_x_at 1437844_x_at 1449053_s_at 1456740_x_at | 14900 | -0.501 | -0.3757 | No | ||
| 46 | ICA1 | 1417901_a_at 1431644_a_at | 15350 | -0.593 | -0.3912 | No | ||
| 47 | MALL | 1425359_at 1435693_at | 15455 | -0.614 | -0.3908 | No | ||
| 48 | A2BP1 | 1418314_a_at 1438217_at 1447735_x_at 1455358_at 1456921_at | 16036 | -0.748 | -0.4110 | No | ||
| 49 | CHST2 | 1422758_at 1460070_at | 16435 | -0.845 | -0.4221 | No | ||
| 50 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 16450 | -0.849 | -0.4155 | No | ||
| 51 | GOLGA4 | 1417674_s_at 1448803_at 1460213_at | 16590 | -0.881 | -0.4144 | No | ||
| 52 | RHOT1 | 1423533_a_at 1440853_at | 16655 | -0.899 | -0.4098 | No | ||
| 53 | COPE | 1416384_a_at | 17283 | -1.034 | -0.4297 | No | ||
| 54 | TAPBP | 1421812_at 1450378_at | 17595 | -1.113 | -0.4345 | No | ||
| 55 | AP4M1 | 1418846_at | 18013 | -1.238 | -0.4431 | Yes | ||
| 56 | ATP7A | 1418774_a_at 1436921_at | 18119 | -1.271 | -0.4372 | Yes | ||
| 57 | GLG1 | 1417087_at 1448579_at 1448580_at 1460554_s_at | 18166 | -1.286 | -0.4284 | Yes | ||
| 58 | ADAM10 | 1450104_at 1460083_at | 18526 | -1.414 | -0.4329 | Yes | ||
| 59 | AP4B1 | 1416631_at 1421726_at | 18570 | -1.434 | -0.4227 | Yes | ||
| 60 | COPA | 1415706_at 1437274_at | 18909 | -1.587 | -0.4247 | Yes | ||
| 61 | TRAPPC4 | 1415674_a_at 1429632_at | 18914 | -1.589 | -0.4115 | Yes | ||
| 62 | COG3 | 1434654_at 1456286_at | 18975 | -1.614 | -0.4005 | Yes | ||
| 63 | OCRL | 1438284_at 1438396_at 1457313_at | 19429 | -1.814 | -0.4059 | Yes | ||
| 64 | B4GALT1 | 1418014_a_at 1458616_at 1458773_at | 19447 | -1.825 | -0.3913 | Yes | ||
| 65 | AP1S1 | 1416087_at | 19580 | -1.906 | -0.3812 | Yes | ||
| 66 | FURIN | 1418518_at | 19626 | -1.944 | -0.3668 | Yes | ||
| 67 | COG2 | 1416932_at | 19898 | -2.164 | -0.3609 | Yes | ||
| 68 | LARGE | 1417435_at 1417436_at 1441642_at 1442420_at 1443212_at 1458663_at | 19917 | -2.184 | -0.3432 | Yes | ||
| 69 | TRIM23 | 1426969_at 1429998_at 1456089_at | 20038 | -2.287 | -0.3294 | Yes | ||
| 70 | AP1G2 | 1419113_at | 20203 | -2.420 | -0.3164 | Yes | ||
| 71 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 20211 | -2.423 | -0.2962 | Yes | ||
| 72 | COG6 | 1426216_at | 20601 | -2.827 | -0.2901 | Yes | ||
| 73 | ZFYVE1 | 1435071_at 1435072_at | 20613 | -2.847 | -0.2665 | Yes | ||
| 74 | COG4 | 1416993_at 1419808_at 1419809_s_at 1443135_at | 20700 | -2.966 | -0.2453 | Yes | ||
| 75 | PCSK7 | 1417180_at | 20917 | -3.310 | -0.2272 | Yes | ||
| 76 | AP1GBP1 | 1435215_at 1442344_at 1457695_at | 21006 | -3.474 | -0.2019 | Yes | ||
| 77 | FIS1 | 1416764_at 1443819_x_at | 21029 | -3.513 | -0.1731 | Yes | ||
| 78 | RHOT2 | 1426822_at | 21256 | -3.972 | -0.1499 | Yes | ||
| 79 | IGF2R | 1424111_at 1424112_at 1440979_at 1445966_at 1457356_at | 21445 | -4.484 | -0.1205 | Yes | ||
| 80 | GGA3 | 1438008_at | 21589 | -5.107 | -0.0839 | Yes | ||
| 81 | DOPEY2 | 1428330_at 1449904_at | 21648 | -5.506 | -0.0399 | Yes | ||
| 82 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 21738 | -6.260 | 0.0090 | Yes |