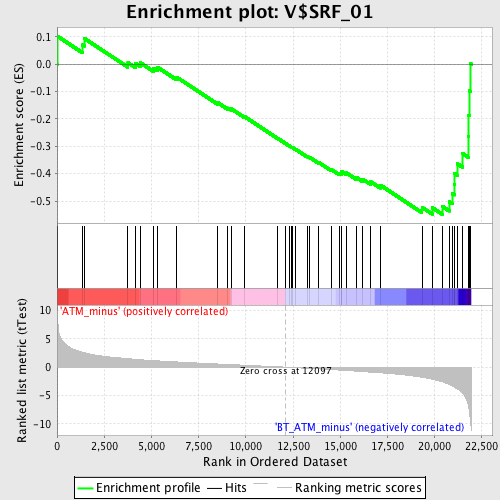
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$SRF_01 |
| Enrichment Score (ES) | -0.54921174 |
| Normalized Enrichment Score (NES) | -1.8546443 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.02257181 |
| FWER p-Value | 0.047 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TPM1 | 1423049_a_at 1423721_at 1447713_at 1456623_at | 5 | 9.273 | 0.1024 | No | ||
| 2 | IGF2AS | 1427394_at | 1349 | 2.629 | 0.0702 | No | ||
| 3 | FOSB | 1422134_at | 1438 | 2.551 | 0.0944 | No | ||
| 4 | GPR20 | 1440021_at | 3751 | 1.495 | 0.0053 | No | ||
| 5 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 4138 | 1.384 | 0.0030 | No | ||
| 6 | ACTR3 | 1426392_a_at 1434968_a_at 1452051_at | 4398 | 1.316 | 0.0057 | No | ||
| 7 | CFL1 | 1448346_at 1455138_x_at | 5119 | 1.138 | -0.0146 | No | ||
| 8 | MRVI1 | 1422245_a_at 1459665_s_at | 5319 | 1.102 | -0.0115 | No | ||
| 9 | PLCB3 | 1448661_at | 6320 | 0.920 | -0.0470 | No | ||
| 10 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 8509 | 0.553 | -0.1408 | No | ||
| 11 | EGR4 | 1449977_at | 9046 | 0.466 | -0.1601 | No | ||
| 12 | PHOX2B | 1422232_at 1455907_x_at | 9216 | 0.440 | -0.1630 | No | ||
| 13 | DIXDC1 | 1425256_a_at 1435207_at 1443957_at 1444395_at | 9904 | 0.337 | -0.1906 | No | ||
| 14 | ASB2 | 1428444_at 1436259_at | 11660 | 0.068 | -0.2700 | No | ||
| 15 | HOXB4 | 1451761_at 1460379_at | 12097 | -0.000 | -0.2900 | No | ||
| 16 | CA3 | 1430584_s_at 1449434_at 1453588_at 1460256_at | 12305 | -0.035 | -0.2990 | No | ||
| 17 | TNNC1 | 1418370_at | 12436 | -0.059 | -0.3043 | No | ||
| 18 | EGR3 | 1421486_at 1436329_at | 12466 | -0.064 | -0.3049 | No | ||
| 19 | IL17B | 1431693_a_at | 12621 | -0.093 | -0.3109 | No | ||
| 20 | FOSL1 | 1417487_at 1417488_at | 13244 | -0.198 | -0.3371 | No | ||
| 21 | KCNMB1 | 1421400_at 1421401_at | 13370 | -0.218 | -0.3404 | No | ||
| 22 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 13834 | -0.297 | -0.3583 | No | ||
| 23 | ITGB1BP2 | 1423238_at | 14508 | -0.421 | -0.3844 | No | ||
| 24 | CNN2 | 1450981_at | 14942 | -0.509 | -0.3985 | No | ||
| 25 | MYH11 | 1418122_at 1448962_at | 15076 | -0.538 | -0.3987 | No | ||
| 26 | SCOC | 1416267_at 1430999_a_at | 15077 | -0.539 | -0.3927 | No | ||
| 27 | VCL | 1416156_at 1416157_at 1445256_at | 15322 | -0.587 | -0.3973 | No | ||
| 28 | MYL6 | 1435041_at | 15833 | -0.697 | -0.4129 | No | ||
| 29 | FOS | 1423100_at | 16185 | -0.784 | -0.4203 | No | ||
| 30 | TNMD | 1417979_at | 16611 | -0.887 | -0.4299 | No | ||
| 31 | PRM1 | 1415955_x_at 1437054_x_at 1439379_x_at | 17111 | -1.001 | -0.4416 | No | ||
| 32 | TGFB1I1 | 1418136_at | 19324 | -1.767 | -0.5231 | No | ||
| 33 | PFN1 | 1449018_at | 19877 | -2.140 | -0.5246 | Yes | ||
| 34 | CAP1 | 1417461_at 1417462_at | 20417 | -2.603 | -0.5204 | Yes | ||
| 35 | IER2 | 1416442_at | 20772 | -3.058 | -0.5027 | Yes | ||
| 36 | ACTG2 | 1422340_a_at | 20925 | -3.331 | -0.4728 | Yes | ||
| 37 | MUS81 | 1431885_a_at 1450432_s_at | 21041 | -3.543 | -0.4388 | Yes | ||
| 38 | TAZ | 1416207_at 1453785_at | 21053 | -3.577 | -0.3997 | Yes | ||
| 39 | FLNA | 1426677_at | 21187 | -3.814 | -0.3636 | Yes | ||
| 40 | WDR1 | 1423054_at 1437591_a_at 1450851_at | 21464 | -4.534 | -0.3260 | Yes | ||
| 41 | JUNB | 1415899_at | 21777 | -6.806 | -0.2649 | Yes | ||
| 42 | EGR1 | 1417065_at | 21803 | -7.184 | -0.1865 | Yes | ||
| 43 | ANXA6 | 1415818_at 1429246_a_at 1429247_at 1442288_at | 21843 | -8.231 | -0.0972 | Yes | ||
| 44 | EGR2 | 1427682_a_at 1427683_at | 21881 | -9.155 | 0.0024 | Yes |

