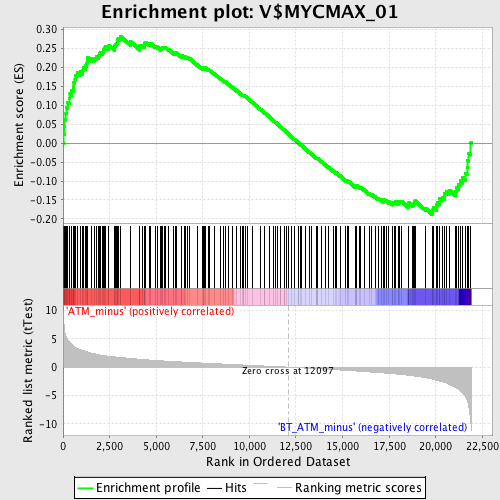
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$MYCMAX_01 |
| Enrichment Score (ES) | 0.2819085 |
| Normalized Enrichment Score (NES) | 1.2658459 |
| Nominal p-value | 0.04589372 |
| FDR q-value | 0.49858493 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SDC1 | 1415943_at 1415944_at 1437279_x_at 1448158_at | 44 | 7.679 | 0.0234 | Yes | ||
| 2 | LYAR | 1417511_at | 78 | 6.311 | 0.0428 | Yes | ||
| 3 | NOL5A | 1426533_at 1455035_s_at | 85 | 6.190 | 0.0630 | Yes | ||
| 4 | LAP3 | 1450860_at | 150 | 5.412 | 0.0780 | Yes | ||
| 5 | PRPS1 | 1416052_at | 182 | 5.217 | 0.0939 | Yes | ||
| 6 | QTRT1 | 1450630_at | 237 | 4.837 | 0.1074 | Yes | ||
| 7 | PRDX4 | 1416166_a_at 1416167_at 1431269_at | 328 | 4.426 | 0.1180 | Yes | ||
| 8 | PUS1 | 1418869_a_at | 367 | 4.281 | 0.1304 | Yes | ||
| 9 | IFRD2 | 1451016_at | 471 | 3.976 | 0.1388 | Yes | ||
| 10 | QTRTD1 | 1421320_a_at 1451199_at | 543 | 3.801 | 0.1482 | Yes | ||
| 11 | RANBP1 | 1422547_at | 561 | 3.755 | 0.1598 | Yes | ||
| 12 | TIMM10 | 1417499_at | 637 | 3.553 | 0.1681 | Yes | ||
| 13 | G6PC3 | 1428245_at | 682 | 3.458 | 0.1776 | Yes | ||
| 14 | ENO3 | 1417951_at 1449631_at | 745 | 3.349 | 0.1858 | Yes | ||
| 15 | TCERG1 | 1421033_a_at 1434434_s_at 1450100_a_at | 909 | 3.083 | 0.1885 | Yes | ||
| 16 | ETV1 | 1422607_at 1445314_at 1450684_at | 1054 | 2.944 | 0.1917 | Yes | ||
| 17 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1090 | 2.905 | 0.1997 | Yes | ||
| 18 | NUDC | 1419471_a_at | 1202 | 2.780 | 0.2038 | Yes | ||
| 19 | NDUFS1 | 1425143_a_at 1453354_at | 1249 | 2.736 | 0.2107 | Yes | ||
| 20 | PPAT | 1428543_at 1441770_at 1452831_s_at | 1290 | 2.688 | 0.2178 | Yes | ||
| 21 | ZADH2 | 1451277_at | 1296 | 2.678 | 0.2264 | Yes | ||
| 22 | PAICS | 1423564_a_at 1423565_at 1436298_x_at | 1548 | 2.452 | 0.2230 | Yes | ||
| 23 | ARMET | 1428112_at 1459455_at | 1695 | 2.342 | 0.2241 | Yes | ||
| 24 | RPS6KA5 | 1431050_at 1437625_at 1439004_at 1440343_at | 1779 | 2.279 | 0.2278 | Yes | ||
| 25 | RCOR2 | 1417302_at | 1897 | 2.194 | 0.2297 | Yes | ||
| 26 | AMPD2 | 1426757_at 1438941_x_at | 1935 | 2.169 | 0.2352 | Yes | ||
| 27 | MYCL1 | 1422087_at 1422088_at 1434777_at | 1991 | 2.134 | 0.2397 | Yes | ||
| 28 | PTGES2 | 1417591_at | 2135 | 2.064 | 0.2400 | Yes | ||
| 29 | FGF6 | 1427582_at | 2161 | 2.043 | 0.2456 | Yes | ||
| 30 | PTMA | 1423455_at | 2198 | 2.027 | 0.2507 | Yes | ||
| 31 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 2272 | 1.990 | 0.2539 | Yes | ||
| 32 | TRIB1 | 1424880_at 1424881_at | 2419 | 1.929 | 0.2536 | Yes | ||
| 33 | SRP72 | 1428876_at 1428877_at | 2462 | 1.913 | 0.2580 | Yes | ||
| 34 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 2753 | 1.806 | 0.2506 | Yes | ||
| 35 | MRPL40 | 1448849_at | 2768 | 1.800 | 0.2560 | Yes | ||
| 36 | HMGA1 | 1416184_s_at | 2813 | 1.783 | 0.2598 | Yes | ||
| 37 | OPRS1 | 1416750_at 1435579_at 1440792_x_at 1451956_a_at 1457324_at | 2876 | 1.761 | 0.2628 | Yes | ||
| 38 | OPRD1 | 1422121_at | 2906 | 1.752 | 0.2673 | Yes | ||
| 39 | H3F3A | 1423263_at 1442087_at | 2927 | 1.744 | 0.2722 | Yes | ||
| 40 | CEBPA | 1418982_at | 2979 | 1.726 | 0.2755 | Yes | ||
| 41 | SUCLG2 | 1427441_a_at 1435841_s_at 1444309_at | 3067 | 1.701 | 0.2772 | Yes | ||
| 42 | BOK | 1417040_a_at | 3087 | 1.695 | 0.2819 | Yes | ||
| 43 | COL2A1 | 1450567_a_at | 3613 | 1.531 | 0.2628 | No | ||
| 44 | TIMM9 | 1428010_at 1449886_a_at | 3630 | 1.526 | 0.2672 | No | ||
| 45 | ACY1 | 1419173_at | 4110 | 1.391 | 0.2498 | No | ||
| 46 | FKBP10 | 1415951_at 1449632_s_at | 4124 | 1.388 | 0.2538 | No | ||
| 47 | HNRPDL | 1420093_s_at 1420094_at 1424251_a_at 1424252_at 1428224_at 1428225_s_at 1449039_a_at 1456698_s_at | 4125 | 1.387 | 0.2583 | No | ||
| 48 | SNX5 | 1417646_a_at 1417647_at 1417648_s_at 1437074_at 1448791_at | 4241 | 1.354 | 0.2575 | No | ||
| 49 | POU3F2 | 1450831_at 1453478_at 1457734_at | 4368 | 1.322 | 0.2561 | No | ||
| 50 | CAD | 1440857_at 1452829_at 1452830_s_at | 4380 | 1.320 | 0.2600 | No | ||
| 51 | SNCAIP | 1423499_at 1430463_a_at 1441037_at | 4391 | 1.317 | 0.2639 | No | ||
| 52 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 4447 | 1.300 | 0.2657 | No | ||
| 53 | RUNX2 | 1424704_at 1425389_a_at 1426034_a_at | 4656 | 1.247 | 0.2602 | No | ||
| 54 | RAI14 | 1417400_at 1417401_at 1441030_at 1444777_at | 4696 | 1.233 | 0.2625 | No | ||
| 55 | ELAVL3 | 1452524_a_at | 4946 | 1.176 | 0.2550 | No | ||
| 56 | DAZL | 1419542_at 1449502_at | 5067 | 1.148 | 0.2533 | No | ||
| 57 | KCTD15 | 1435339_at | 5251 | 1.114 | 0.2486 | No | ||
| 58 | PCDHA10 | 1427687_at | 5309 | 1.103 | 0.2496 | No | ||
| 59 | TCOF1 | 1423600_a_at 1423601_s_at 1424643_at | 5335 | 1.098 | 0.2521 | No | ||
| 60 | MXD4 | 1422574_at 1434378_a_at 1434379_at | 5421 | 1.082 | 0.2518 | No | ||
| 61 | SFXN2 | 1439966_x_at 1440786_x_at | 5489 | 1.069 | 0.2522 | No | ||
| 62 | FKBP11 | 1417267_s_at 1425839_at 1425938_a_at | 5680 | 1.030 | 0.2469 | No | ||
| 63 | HOXB5 | 1418415_at | 5953 | 0.981 | 0.2376 | No | ||
| 64 | TFRC | 1422966_a_at 1422967_a_at 1452661_at AFFX-TransRecMur/X57349_3_at AFFX-TransRecMur/X57349_5_at AFFX-TransRecMur/X57349_M_at | 6035 | 0.966 | 0.2371 | No | ||
| 65 | SMAD2 | 1420634_a_at 1458304_at | 6070 | 0.961 | 0.2387 | No | ||
| 66 | GIT1 | 1454759_at | 6361 | 0.914 | 0.2284 | No | ||
| 67 | GPM6B | 1423091_a_at 1425942_a_at 1453514_at | 6368 | 0.912 | 0.2312 | No | ||
| 68 | BTBD5 | 1428721_at 1453987_at | 6523 | 0.881 | 0.2270 | No | ||
| 69 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 6583 | 0.870 | 0.2272 | No | ||
| 70 | ETV4 | 1423232_at 1443381_at | 6704 | 0.849 | 0.2245 | No | ||
| 71 | DIABLO | 1417683_at 1425768_at | 6777 | 0.835 | 0.2239 | No | ||
| 72 | SYT3 | 1417708_at | 7199 | 0.763 | 0.2071 | No | ||
| 73 | APOA5 | 1417610_at | 7478 | 0.719 | 0.1967 | No | ||
| 74 | GADD45G | 1453851_a_at | 7540 | 0.708 | 0.1963 | No | ||
| 75 | RHEBL1 | 1447818_x_at 1451516_at | 7543 | 0.708 | 0.1985 | No | ||
| 76 | KIAA0427 | 1427743_at 1436047_at | 7572 | 0.703 | 0.1996 | No | ||
| 77 | SLC12A5 | 1425337_at 1445933_at 1451674_at | 7630 | 0.694 | 0.1992 | No | ||
| 78 | FLT3 | 1419538_at | 7780 | 0.671 | 0.1946 | No | ||
| 79 | SCRT2 | 1440930_a_at 1440931_at 1446986_at 1457310_x_at | 7877 | 0.652 | 0.1924 | No | ||
| 80 | SATB2 | 1425904_at 1427017_at | 8118 | 0.613 | 0.1833 | No | ||
| 81 | FOXD3 | 1422210_at | 8439 | 0.564 | 0.1705 | No | ||
| 82 | RTN4RL2 | 1439573_at | 8616 | 0.534 | 0.1642 | No | ||
| 83 | LIN28 | 1437752_at | 8700 | 0.521 | 0.1621 | No | ||
| 84 | SLC6A15 | 1426712_at | 8899 | 0.488 | 0.1546 | No | ||
| 85 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 9079 | 0.461 | 0.1479 | No | ||
| 86 | NR6A1 | 1421515_at 1421516_at 1428825_at 1428826_at 1445872_at | 9336 | 0.423 | 0.1375 | No | ||
| 87 | ZBTB10 | 1458695_at | 9533 | 0.395 | 0.1298 | No | ||
| 88 | TUBA4 | 1417373_a_at 1417374_at 1417375_at | 9651 | 0.379 | 0.1257 | No | ||
| 89 | RPS19 | 1449243_a_at 1460442_at | 9687 | 0.373 | 0.1253 | No | ||
| 90 | IVNS1ABP | 1420961_a_at 1425718_a_at 1450084_s_at | 9710 | 0.368 | 0.1255 | No | ||
| 91 | KCNQ5 | 1427426_at 1441071_at | 9790 | 0.354 | 0.1231 | No | ||
| 92 | ATAD3A | 1422461_at 1437343_x_at 1438178_x_at 1456541_x_at | 9882 | 0.340 | 0.1200 | No | ||
| 93 | PPARGC1B | 1438775_at 1449945_at 1457578_at | 10187 | 0.294 | 0.1070 | No | ||
| 94 | GPRC5C | 1424450_at 1452947_at | 10600 | 0.231 | 0.0888 | No | ||
| 95 | NEUROD1 | 1426412_at 1426413_at | 10817 | 0.197 | 0.0796 | No | ||
| 96 | USP15 | 1436891_at 1446186_at 1454036_a_at 1455842_x_at | 10832 | 0.195 | 0.0796 | No | ||
| 97 | DNAJB5 | 1421961_a_at 1421962_at 1450436_s_at | 11062 | 0.161 | 0.0696 | No | ||
| 98 | EIF2C2 | 1426366_at 1438043_at | 11313 | 0.121 | 0.0585 | No | ||
| 99 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 11415 | 0.107 | 0.0542 | No | ||
| 100 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 11494 | 0.095 | 0.0509 | No | ||
| 101 | EFNB1 | 1418285_at 1418286_a_at 1451591_a_at | 11686 | 0.064 | 0.0424 | No | ||
| 102 | PPRC1 | 1426381_at | 11697 | 0.062 | 0.0421 | No | ||
| 103 | PSME3 | 1418078_at 1418079_at 1438509_at | 11864 | 0.036 | 0.0346 | No | ||
| 104 | HOXA11 | 1420414_at | 11866 | 0.035 | 0.0347 | No | ||
| 105 | EEF1B2 | 1448252_a_at | 11973 | 0.019 | 0.0298 | No | ||
| 106 | EIF4E | 1450909_at 1457489_at 1459371_at | 12117 | -0.004 | 0.0233 | No | ||
| 107 | STXBP2 | 1416449_x_at 1437048_x_at 1448341_a_at | 12251 | -0.024 | 0.0172 | No | ||
| 108 | DAZAP1 | 1419275_at | 12427 | -0.056 | 0.0094 | No | ||
| 109 | UBE2B | 1423106_at 1423107_at 1430177_at 1439477_at 1460132_at | 12633 | -0.095 | 0.0003 | No | ||
| 110 | MEOX2 | 1424233_at 1424234_s_at | 12662 | -0.101 | -0.0007 | No | ||
| 111 | STMN1 | 1415849_s_at | 12732 | -0.110 | -0.0035 | No | ||
| 112 | NRIP3 | 1418104_at 1431597_a_at 1448954_at | 12822 | -0.126 | -0.0072 | No | ||
| 113 | NRAS | 1422688_a_at 1454060_a_at | 13011 | -0.158 | -0.0153 | No | ||
| 114 | KCMF1 | 1418694_at 1418695_a_at 1449180_at | 13204 | -0.190 | -0.0235 | No | ||
| 115 | CSK | 1423518_at 1439744_at | 13313 | -0.210 | -0.0277 | No | ||
| 116 | CLN3 | 1417551_at 1457789_at | 13611 | -0.260 | -0.0405 | No | ||
| 117 | PITX3 | 1449917_at | 13617 | -0.262 | -0.0399 | No | ||
| 118 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 13645 | -0.267 | -0.0403 | No | ||
| 119 | ATOH8 | 1426418_at | 13665 | -0.271 | -0.0402 | No | ||
| 120 | USP34 | 1434392_at 1434393_at 1444437_at 1446112_at 1460126_at | 13862 | -0.301 | -0.0482 | No | ||
| 121 | EWSR1 | 1417238_at 1425919_at 1436884_x_at 1441074_at 1456461_at | 14064 | -0.336 | -0.0564 | No | ||
| 122 | FGF14 | 1435747_at 1447221_x_at 1447480_at 1449958_a_at 1458830_at 1459073_x_at | 14229 | -0.367 | -0.0627 | No | ||
| 123 | ADAMTS19 | 1442459_at 1446513_at | 14493 | -0.417 | -0.0734 | No | ||
| 124 | PPP1R9B | 1447247_at 1447486_at | 14644 | -0.447 | -0.0788 | No | ||
| 125 | GJA1 | 1415800_at 1415801_at 1437992_x_at 1438650_x_at 1438945_x_at 1438973_x_at | 14663 | -0.450 | -0.0782 | No | ||
| 126 | ABCA1 | 1421839_at 1421840_at 1450392_at | 14875 | -0.494 | -0.0862 | No | ||
| 127 | CHRM1 | 1439611_at 1450833_at | 15181 | -0.558 | -0.0984 | No | ||
| 128 | HNRPA1 | 1423531_a_at 1430020_x_at 1436549_a_at 1442758_at 1446038_at 1455305_x_at 1455740_at | 15253 | -0.575 | -0.0998 | No | ||
| 129 | MPP3 | 1419077_at 1459611_at | 15338 | -0.591 | -0.1017 | No | ||
| 130 | HHIP | 1421426_at 1437933_at 1438083_at 1455277_at | 15690 | -0.671 | -0.1156 | No | ||
| 131 | KRTCAP2 | 1417058_a_at 1417059_at | 15693 | -0.671 | -0.1134 | No | ||
| 132 | IPO7 | 1442511_at 1454955_at 1458165_at | 15761 | -0.683 | -0.1143 | No | ||
| 133 | MNT | 1418192_at | 15763 | -0.683 | -0.1120 | No | ||
| 134 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 15919 | -0.719 | -0.1168 | No | ||
| 135 | UBXD3 | 1426007_a_at 1451906_at 1456385_x_at 1457581_at | 15969 | -0.731 | -0.1166 | No | ||
| 136 | NTN2L | 1422056_at | 16172 | -0.781 | -0.1233 | No | ||
| 137 | RFX4 | 1432053_at 1436931_at 1443312_at | 16440 | -0.846 | -0.1328 | No | ||
| 138 | RAB3IL1 | 1419991_at 1419992_x_at 1419993_at 1425133_s_at 1428391_at 1456442_at | 16570 | -0.875 | -0.1358 | No | ||
| 139 | SLC1A7 | 1456971_at | 16764 | -0.929 | -0.1416 | No | ||
| 140 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 16915 | -0.960 | -0.1453 | No | ||
| 141 | POGK | 1447864_s_at 1459896_at | 17082 | -0.995 | -0.1497 | No | ||
| 142 | MAT2A | 1423667_at 1433576_at 1438386_x_at 1438630_x_at 1438976_x_at 1439386_x_at 1456702_x_at | 17211 | -1.021 | -0.1522 | No | ||
| 143 | BMP4 | 1422912_at | 17235 | -1.026 | -0.1498 | No | ||
| 144 | RFX5 | 1423103_at | 17346 | -1.046 | -0.1514 | No | ||
| 145 | JMJD1A | 1426810_at | 17499 | -1.090 | -0.1548 | No | ||
| 146 | TRIM46 | 1460568_at | 17684 | -1.135 | -0.1595 | No | ||
| 147 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 17694 | -1.138 | -0.1561 | No | ||
| 148 | BCL7C | 1440906_at 1443898_at 1446516_at 1447139_at 1449523_at | 17772 | -1.162 | -0.1558 | No | ||
| 149 | SFRS14 | 1435798_a_at 1435799_at 1436005_at | 17823 | -1.179 | -0.1542 | No | ||
| 150 | ILF3 | 1422546_at 1436802_at 1460669_at | 17990 | -1.232 | -0.1578 | No | ||
| 151 | ARL3 | 1450706_a_at | 17991 | -1.232 | -0.1537 | No | ||
| 152 | GPS1 | 1415699_a_at | 18090 | -1.265 | -0.1540 | No | ||
| 153 | RAMP2 | 1418187_at | 18162 | -1.284 | -0.1530 | No | ||
| 154 | U2AF2 | 1417260_at | 18540 | -1.421 | -0.1656 | No | ||
| 155 | IPO13 | 1433464_at | 18541 | -1.421 | -0.1609 | No | ||
| 156 | HIRA | 1436241_s_at 1450049_a_at 1450050_at 1459103_at | 18571 | -1.435 | -0.1575 | No | ||
| 157 | HSPBAP1 | 1428566_at 1428567_at 1441900_x_at 1442842_at | 18772 | -1.525 | -0.1617 | No | ||
| 158 | DIRC2 | 1454654_at 1457245_at | 18840 | -1.552 | -0.1596 | No | ||
| 159 | ASXL1 | 1434561_at 1435077_at 1458380_at | 18852 | -1.561 | -0.1549 | No | ||
| 160 | ZHX2 | 1455210_at | 18906 | -1.585 | -0.1521 | No | ||
| 161 | CNNM1 | 1420354_at 1441312_at 1445979_at | 19457 | -1.834 | -0.1713 | No | ||
| 162 | FOXO3A | 1434831_a_at 1434832_at 1439598_at 1444226_at 1450236_at 1458279_at | 19826 | -2.098 | -0.1813 | No | ||
| 163 | CBX5 | 1421932_at 1421933_at 1421934_at 1450416_at 1454636_at 1459845_at | 19860 | -2.130 | -0.1758 | No | ||
| 164 | PFN1 | 1449018_at | 19877 | -2.140 | -0.1694 | No | ||
| 165 | PER1 | 1449851_at | 20057 | -2.301 | -0.1700 | No | ||
| 166 | ESRRA | 1442864_at 1460652_at | 20062 | -2.306 | -0.1626 | No | ||
| 167 | CGREF1 | 1424528_at 1424529_s_at | 20101 | -2.343 | -0.1565 | No | ||
| 168 | WDR23 | 1448418_s_at 1460189_at | 20220 | -2.429 | -0.1539 | No | ||
| 169 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 20221 | -2.430 | -0.1459 | No | ||
| 170 | FOXJ3 | 1435362_at 1438220_at 1459055_at | 20371 | -2.562 | -0.1442 | No | ||
| 171 | LZTS2 | 1426409_at | 20464 | -2.654 | -0.1397 | No | ||
| 172 | BCAS3 | 1423528_at 1428454_at 1457817_at 1458557_at | 20484 | -2.677 | -0.1317 | No | ||
| 173 | TESK2 | 1426546_at | 20596 | -2.819 | -0.1274 | No | ||
| 174 | KCNN4 | 1421038_a_at 1435945_a_at | 20748 | -3.034 | -0.1243 | No | ||
| 175 | CRMP1 | 1448289_at | 21063 | -3.593 | -0.1269 | No | ||
| 176 | MTHFD1 | 1415916_a_at 1415917_at 1436704_x_at | 21137 | -3.732 | -0.1178 | No | ||
| 177 | GPC3 | 1443240_at 1446525_at 1450990_at | 21242 | -3.939 | -0.1096 | No | ||
| 178 | UBE1 | 1442767_s_at 1448116_at | 21315 | -4.103 | -0.0993 | No | ||
| 179 | IQGAP2 | 1433885_at 1459894_at | 21449 | -4.503 | -0.0905 | No | ||
| 180 | RORC | 1425792_a_at 1425793_a_at | 21623 | -5.306 | -0.0809 | No | ||
| 181 | SLC43A1 | 1440007_at 1453255_at | 21694 | -5.787 | -0.0649 | No | ||
| 182 | TUBA1 | 1418884_x_at | 21709 | -5.969 | -0.0458 | No | ||
| 183 | ISGF3G | 1421322_a_at | 21788 | -6.962 | -0.0263 | No | ||
| 184 | RCL1 | 1416730_at 1457992_at | 21901 | -9.952 | 0.0015 | No |

