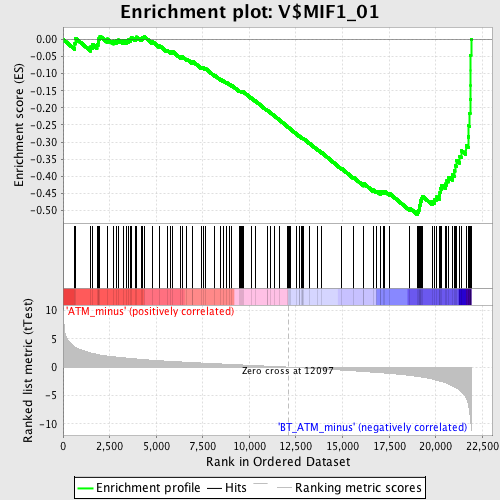
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$MIF1_01 |
| Enrichment Score (ES) | -0.5118769 |
| Normalized Enrichment Score (NES) | -2.0223286 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0034907712 |
| FWER p-Value | 0.0050 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHCHD4 | 1417233_at | 631 | 3.563 | -0.0117 | No | ||
| 2 | GRID2 | 1421435_at 1421436_at 1435487_at 1437824_at 1443101_at 1446879_at 1458756_at 1459245_s_at | 664 | 3.496 | 0.0037 | No | ||
| 3 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 1485 | 2.505 | -0.0218 | No | ||
| 4 | HAT1 | 1428061_at | 1588 | 2.416 | -0.0148 | No | ||
| 5 | NEK2 | 1417299_at 1437579_at 1437580_s_at 1443999_at | 1820 | 2.250 | -0.0145 | No | ||
| 6 | FIBCD1 | 1435482_at | 1877 | 2.213 | -0.0064 | No | ||
| 7 | SOX3 | 1435192_at 1450485_at | 1921 | 2.177 | 0.0021 | No | ||
| 8 | CRLF1 | 1418476_at | 1978 | 2.142 | 0.0099 | No | ||
| 9 | SMAD5 | 1421047_at 1433641_at 1451873_a_at | 2366 | 1.950 | 0.0016 | No | ||
| 10 | CARS | 1427330_at 1452394_at | 2689 | 1.828 | -0.0043 | No | ||
| 11 | TLX3 | 1450806_at | 2849 | 1.768 | -0.0031 | No | ||
| 12 | FGR | 1419526_at 1442804_at | 2992 | 1.723 | -0.0013 | No | ||
| 13 | GRIK5 | 1418784_at | 3219 | 1.656 | -0.0036 | No | ||
| 14 | PHF15 | 1430081_at 1455345_at | 3389 | 1.604 | -0.0036 | No | ||
| 15 | PTCH2 | 1422655_at | 3498 | 1.565 | -0.0010 | No | ||
| 16 | MDH1B | 1429842_at | 3604 | 1.534 | 0.0016 | No | ||
| 17 | TUSC3 | 1421662_a_at 1458568_at | 3647 | 1.521 | 0.0070 | No | ||
| 18 | GALNT14 | 1453630_at | 3889 | 1.455 | 0.0030 | No | ||
| 19 | EMX2 | 1456258_at | 3945 | 1.442 | 0.0074 | No | ||
| 20 | TPCN1 | 1434930_at 1451771_at 1451772_at | 4200 | 1.366 | 0.0023 | No | ||
| 21 | TBC1D16 | 1435763_at 1442149_at | 4273 | 1.348 | 0.0056 | No | ||
| 22 | PLA2G12A | 1452026_a_at | 4343 | 1.330 | 0.0088 | No | ||
| 23 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 4805 | 1.206 | -0.0065 | No | ||
| 24 | MYCBP | 1421165_at 1427938_at 1427939_s_at 1427940_s_at 1452608_at | 5191 | 1.126 | -0.0187 | No | ||
| 25 | BMP6 | 1450759_at 1459947_at | 5581 | 1.051 | -0.0314 | No | ||
| 26 | PTDSS2 | 1426728_x_at 1429492_x_at 1434586_a_at 1434587_x_at 1439984_at 1443738_at 1450354_a_at 1453164_a_at 1458387_at 1460010_a_at | 5788 | 1.011 | -0.0360 | No | ||
| 27 | LRP2 | 1427133_s_at 1427621_at 1445901_at 1452320_at | 5895 | 0.993 | -0.0361 | No | ||
| 28 | PLCB3 | 1448661_at | 6320 | 0.920 | -0.0511 | No | ||
| 29 | TTC16 | 1455414_at | 6406 | 0.903 | -0.0506 | No | ||
| 30 | TEKT2 | 1418484_at | 6640 | 0.860 | -0.0571 | No | ||
| 31 | GRM3 | 1430136_at | 6939 | 0.810 | -0.0669 | No | ||
| 32 | GLRA3 | 1450239_at | 6965 | 0.804 | -0.0641 | No | ||
| 33 | CSMD3 | 1438272_at 1438572_at 1454245_at | 7445 | 0.723 | -0.0826 | No | ||
| 34 | SDCBP2 | 1424090_at | 7520 | 0.712 | -0.0825 | No | ||
| 35 | TTC8 | 1424410_at | 7648 | 0.691 | -0.0850 | No | ||
| 36 | CPEB2 | 1434272_at 1443017_at 1458518_at | 8152 | 0.608 | -0.1051 | No | ||
| 37 | WARS2 | 1429279_at 1435273_at | 8476 | 0.558 | -0.1173 | No | ||
| 38 | NAT5 | 1418244_at 1422961_at | 8618 | 0.534 | -0.1211 | No | ||
| 39 | LSM7 | 1417313_at | 8776 | 0.510 | -0.1259 | No | ||
| 40 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 8935 | 0.483 | -0.1308 | No | ||
| 41 | TGFB3 | 1417455_at | 9064 | 0.463 | -0.1344 | No | ||
| 42 | NRGN | 1423231_at | 9473 | 0.404 | -0.1512 | No | ||
| 43 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 9520 | 0.398 | -0.1513 | No | ||
| 44 | DNAH9 | 1442412_at 1457401_at | 9601 | 0.386 | -0.1531 | No | ||
| 45 | HM13 | 1417287_at 1428855_at 1428856_at 1438456_at 1438865_at 1455631_at | 9627 | 0.382 | -0.1525 | No | ||
| 46 | PACRG | 1428751_at | 9663 | 0.377 | -0.1522 | No | ||
| 47 | TBPL1 | 1431415_a_at 1434907_at | 10104 | 0.307 | -0.1709 | No | ||
| 48 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 10304 | 0.273 | -0.1787 | No | ||
| 49 | PRKCSH | 1416339_a_at | 10950 | 0.178 | -0.2074 | No | ||
| 50 | HMGCS1 | 1433443_a_at 1433444_at 1433445_x_at 1433446_at 1441536_at | 10973 | 0.174 | -0.2076 | No | ||
| 51 | FILIP1 | 1436650_at 1444075_at | 11150 | 0.146 | -0.2149 | No | ||
| 52 | MAGED1 | 1450062_a_at | 11368 | 0.113 | -0.2243 | No | ||
| 53 | ARL4A | 1425411_at 1431429_a_at 1435092_at | 11622 | 0.074 | -0.2356 | No | ||
| 54 | NEDD8 | 1423715_a_at 1440996_at 1445811_at | 12044 | 0.008 | -0.2548 | No | ||
| 55 | STMN4 | 1418105_at | 12122 | -0.005 | -0.2583 | No | ||
| 56 | MRPL10 | 1418112_at | 12173 | -0.012 | -0.2606 | No | ||
| 57 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 12203 | -0.016 | -0.2618 | No | ||
| 58 | BBS2 | 1424478_at | 12506 | -0.072 | -0.2753 | No | ||
| 59 | SPA17 | 1417635_at | 12667 | -0.101 | -0.2822 | No | ||
| 60 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 12691 | -0.104 | -0.2827 | No | ||
| 61 | GPRC5D | 1420538_at | 12827 | -0.126 | -0.2883 | No | ||
| 62 | ITGB8 | 1436223_at | 12842 | -0.129 | -0.2883 | No | ||
| 63 | SALL2 | 1416638_at | 12872 | -0.134 | -0.2890 | No | ||
| 64 | EFNB3 | 1423085_at | 12920 | -0.144 | -0.2904 | No | ||
| 65 | SNF1LK | 1419766_at 1459601_at | 13237 | -0.196 | -0.3040 | No | ||
| 66 | NELF | 1436959_x_at 1439398_x_at 1442706_at 1449474_a_at | 13672 | -0.272 | -0.3226 | No | ||
| 67 | ARID1A | 1447104_at | 13897 | -0.307 | -0.3313 | No | ||
| 68 | MDH1 | 1434319_at 1436834_x_at 1438338_at 1448172_at 1454925_x_at | 14958 | -0.514 | -0.3774 | No | ||
| 69 | RNF41 | 1423842_a_at 1432003_a_at 1453633_a_at 1455763_at | 15591 | -0.648 | -0.4033 | No | ||
| 70 | GTF3C4 | 1434320_at 1438692_at | 16144 | -0.774 | -0.4248 | No | ||
| 71 | WWP1 | 1427097_at 1427098_at 1446364_at 1446853_at 1452299_at | 16145 | -0.774 | -0.4211 | No | ||
| 72 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 16654 | -0.899 | -0.4400 | No | ||
| 73 | THNSL1 | 1427936_at 1452605_at | 16848 | -0.947 | -0.4443 | No | ||
| 74 | CBX6 | 1424407_s_at 1429290_at | 17037 | -0.985 | -0.4482 | No | ||
| 75 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 17050 | -0.989 | -0.4439 | No | ||
| 76 | NEDD9 | 1422818_at 1437132_x_at 1447885_x_at 1450767_at | 17215 | -1.022 | -0.4465 | No | ||
| 77 | NCDN | 1416852_a_at 1416853_at | 17269 | -1.032 | -0.4440 | No | ||
| 78 | RASGEF1A | 1429316_at 1435218_at | 17534 | -1.099 | -0.4508 | No | ||
| 79 | VEGF | 1420909_at 1451959_a_at | 18611 | -1.453 | -0.4930 | No | ||
| 80 | GMPR2 | 1416356_at 1429180_at | 19023 | -1.628 | -0.5040 | Yes | ||
| 81 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 19093 | -1.661 | -0.4992 | Yes | ||
| 82 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 19118 | -1.672 | -0.4922 | Yes | ||
| 83 | ADCK4 | 1448490_at | 19144 | -1.683 | -0.4852 | Yes | ||
| 84 | PARK2 | 1420755_a_at 1426135_a_at 1449975_a_at | 19174 | -1.694 | -0.4784 | Yes | ||
| 85 | KIF17 | 1419826_at 1419827_s_at 1422762_at | 19208 | -1.708 | -0.4716 | Yes | ||
| 86 | POLD4 | 1427885_at | 19246 | -1.730 | -0.4650 | Yes | ||
| 87 | DPAGT1 | 1448549_a_at | 19275 | -1.744 | -0.4578 | Yes | ||
| 88 | TRIM39 | 1418851_at | 19831 | -2.106 | -0.4731 | Yes | ||
| 89 | KIF3B | 1420987_at 1450073_at 1450074_at | 19961 | -2.219 | -0.4683 | Yes | ||
| 90 | BDNF | 1422168_a_at 1422169_a_at | 20032 | -2.284 | -0.4605 | Yes | ||
| 91 | DCTN1 | 1422521_at 1435414_s_at | 20197 | -2.416 | -0.4563 | Yes | ||
| 92 | STK36 | 1434733_at 1453584_at | 20234 | -2.441 | -0.4462 | Yes | ||
| 93 | DNAJC1 | 1420500_at 1420501_at 1440551_at 1447649_x_at 1453372_at 1459791_at | 20269 | -2.465 | -0.4359 | Yes | ||
| 94 | FGF13 | 1418497_at 1418498_at | 20312 | -2.506 | -0.4257 | Yes | ||
| 95 | E2F5 | 1417444_at 1447625_at 1460207_s_at | 20534 | -2.745 | -0.4226 | Yes | ||
| 96 | AP1M1 | 1416307_at 1459832_s_at | 20609 | -2.842 | -0.4122 | Yes | ||
| 97 | CBLB | 1437304_at 1455082_at 1458469_at | 20711 | -2.984 | -0.4025 | Yes | ||
| 98 | SREBF1 | 1426690_a_at | 20907 | -3.287 | -0.3955 | Yes | ||
| 99 | NCOA5 | 1424055_at 1456997_at | 21014 | -3.483 | -0.3836 | Yes | ||
| 100 | RFX2 | 1417332_at | 21045 | -3.551 | -0.3678 | Yes | ||
| 101 | PSCD2 | 1450103_a_at | 21138 | -3.734 | -0.3540 | Yes | ||
| 102 | TMEM24 | 1428095_a_at 1457585_at | 21304 | -4.084 | -0.3418 | Yes | ||
| 103 | MADD | 1425735_at 1455502_at | 21387 | -4.284 | -0.3249 | Yes | ||
| 104 | SPRY2 | 1421656_at 1436584_at | 21635 | -5.415 | -0.3101 | Yes | ||
| 105 | DDX31 | 1434182_at 1438962_s_at | 21757 | -6.438 | -0.2846 | Yes | ||
| 106 | CBX4 | 1419583_at 1440479_at | 21780 | -6.886 | -0.2523 | Yes | ||
| 107 | SOCS1 | 1440047_at 1450446_a_at | 21837 | -8.094 | -0.2158 | Yes | ||
| 108 | CCRK | 1422494_s_at | 21854 | -8.448 | -0.1758 | Yes | ||
| 109 | DHRS3 | 1448390_a_at | 21875 | -8.975 | -0.1334 | Yes | ||
| 110 | RNF25 | 1442108_at 1448674_at | 21879 | -9.080 | -0.0897 | Yes | ||
| 111 | CCDC6 | 1428311_at 1459159_a_at | 21880 | -9.114 | -0.0457 | Yes | ||
| 112 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21904 | -9.965 | 0.0014 | Yes |

