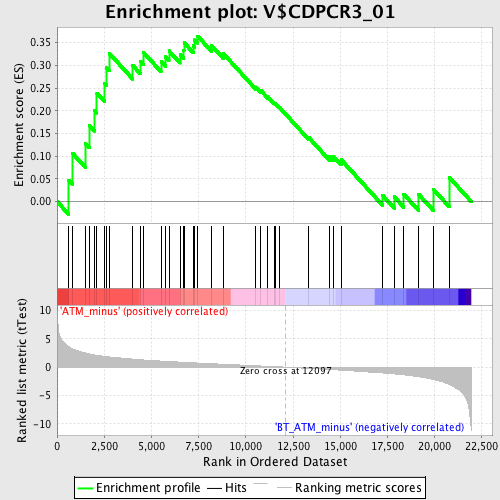
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$CDPCR3_01 |
| Enrichment Score (ES) | 0.36419368 |
| Normalized Enrichment Score (NES) | 1.2430959 |
| Nominal p-value | 0.14498934 |
| FDR q-value | 0.49898154 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HES1 | 1418102_at | 621 | 3.588 | 0.0475 | Yes | ||
| 2 | PTGFRN | 1421319_at 1434891_at | 827 | 3.203 | 0.1059 | Yes | ||
| 3 | NPAS1 | 1420801_at | 1497 | 2.497 | 0.1281 | Yes | ||
| 4 | CDK2AP1 | 1426885_a_at 1435509_x_at 1456314_x_at | 1709 | 2.331 | 0.1678 | Yes | ||
| 5 | SIX1 | 1427277_at | 1985 | 2.139 | 0.2005 | Yes | ||
| 6 | PPARG | 1420715_a_at | 2109 | 2.076 | 0.2387 | Yes | ||
| 7 | FOXB1 | 1420705_at | 2508 | 1.895 | 0.2606 | Yes | ||
| 8 | IRAK1 | 1448668_a_at 1460649_at | 2627 | 1.850 | 0.2944 | Yes | ||
| 9 | ZIC4 | 1421539_at 1456417_at | 2764 | 1.802 | 0.3263 | Yes | ||
| 10 | SIX3 | 1426637_a_at 1426638_at | 4005 | 1.421 | 0.2997 | Yes | ||
| 11 | UPP2 | 1460059_at | 4416 | 1.311 | 0.3087 | Yes | ||
| 12 | L3MBTL2 | 1433777_at | 4567 | 1.266 | 0.3286 | Yes | ||
| 13 | FGF10 | 1420690_at | 5504 | 1.065 | 0.3084 | Yes | ||
| 14 | MYT1 | 1422773_at 1439365_at | 5754 | 1.017 | 0.3185 | Yes | ||
| 15 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 5926 | 0.987 | 0.3316 | Yes | ||
| 16 | FGF7 | 1422243_at 1438405_at | 6509 | 0.883 | 0.3237 | Yes | ||
| 17 | CER1 | 1450256_at 1450257_at | 6701 | 0.849 | 0.3329 | Yes | ||
| 18 | MARK1 | 1419834_x_at 1425509_at 1425510_at 1425511_at 1449630_s_at | 6722 | 0.846 | 0.3499 | Yes | ||
| 19 | LPHN2 | 1434111_at 1434112_at 1444906_at | 7222 | 0.760 | 0.3432 | Yes | ||
| 20 | RBPMS | 1425652_s_at 1429359_s_at 1437161_x_at 1444765_at 1455936_a_at 1459078_at | 7294 | 0.749 | 0.3558 | Yes | ||
| 21 | CSMD3 | 1438272_at 1438572_at 1454245_at | 7445 | 0.723 | 0.3642 | Yes | ||
| 22 | TCF1 | 1421234_at | 8178 | 0.604 | 0.3435 | No | ||
| 23 | PAK3 | 1417923_at 1417924_at 1437318_at | 8795 | 0.506 | 0.3261 | No | ||
| 24 | DLX5 | 1449863_a_at | 10525 | 0.240 | 0.2522 | No | ||
| 25 | ZIC1 | 1423477_at 1439627_at | 10790 | 0.201 | 0.2444 | No | ||
| 26 | CNTN6 | 1422649_at | 11166 | 0.143 | 0.2303 | No | ||
| 27 | MICAL2 | 1438726_at 1455685_at | 11507 | 0.093 | 0.2167 | No | ||
| 28 | CRH | 1457984_at | 11569 | 0.085 | 0.2158 | No | ||
| 29 | HOXA4 | 1420226_x_at 1427354_at | 11784 | 0.048 | 0.2070 | No | ||
| 30 | NFIA | 1421162_a_at 1421163_a_at 1427733_a_at 1438236_at 1440660_at 1441547_at 1446742_at 1446990_at 1456087_at 1459431_at | 13333 | -0.212 | 0.1408 | No | ||
| 31 | ETV3 | 1418635_at 1418636_at 1445997_at 1456358_at | 14438 | -0.407 | 0.0990 | No | ||
| 32 | HDAC4 | 1436758_at 1447566_at 1454693_at | 14637 | -0.445 | 0.0994 | No | ||
| 33 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 15046 | -0.533 | 0.0920 | No | ||
| 34 | BMP4 | 1422912_at | 17235 | -1.026 | 0.0138 | No | ||
| 35 | POU3F4 | 1422164_at 1422165_at | 17853 | -1.188 | 0.0107 | No | ||
| 36 | BHLHB3 | 1421099_at | 18362 | -1.354 | 0.0161 | No | ||
| 37 | SPP2 | 1418916_a_at 1446777_at | 19142 | -1.683 | 0.0162 | No | ||
| 38 | NTN1 | 1422987_at 1454974_at | 19946 | -2.204 | 0.0261 | No | ||
| 39 | PIP5K1A | 1421833_at 1421834_at 1450389_s_at | 20773 | -3.058 | 0.0530 | No |

