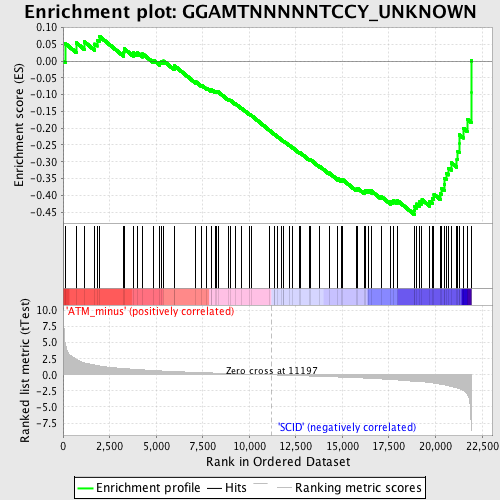
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | GGAMTNNNNNTCCY_UNKNOWN |
| Enrichment Score (ES) | -0.45711708 |
| Normalized Enrichment Score (NES) | -1.6905936 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.3117413 |
| FWER p-Value | 0.214 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACP6 | 1444410_at 1448445_at | 120 | 4.719 | 0.0523 | No | ||
| 2 | HIRIP3 | 1434936_at | 704 | 2.383 | 0.0548 | No | ||
| 3 | CSNK2B | 1416728_at | 1137 | 1.851 | 0.0576 | No | ||
| 4 | NAPB | 1423172_at 1423173_at 1427470_s_at 1450888_at 1452444_at | 1689 | 1.515 | 0.0510 | No | ||
| 5 | FHIT | 1425893_a_at | 1822 | 1.446 | 0.0626 | No | ||
| 6 | FOXA3 | 1431900_a_at | 1936 | 1.387 | 0.0744 | No | ||
| 7 | TPR | 1426948_at 1426949_s_at 1456112_at 1456651_a_at | 3262 | 0.972 | 0.0257 | No | ||
| 8 | UGCGL2 | 1429836_at 1431602_a_at | 3270 | 0.970 | 0.0373 | No | ||
| 9 | PRKCI | 1417410_s_at 1448695_at | 3767 | 0.864 | 0.0251 | No | ||
| 10 | KLHDC3 | 1415991_a_at 1448164_at 1454747_a_at 1455910_at | 3982 | 0.823 | 0.0254 | No | ||
| 11 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 4275 | 0.769 | 0.0215 | No | ||
| 12 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 4852 | 0.666 | 0.0033 | No | ||
| 13 | RRAS | 1418448_at | 5195 | 0.608 | -0.0050 | No | ||
| 14 | CNOT4 | 1420999_at 1421000_at 1436645_a_at 1437586_at 1443169_at 1450083_at 1459449_at | 5266 | 0.598 | -0.0008 | No | ||
| 15 | BRMS1L | 1452850_s_at 1460443_at | 5396 | 0.581 | 0.0004 | No | ||
| 16 | RPIA | 1418337_at | 5965 | 0.505 | -0.0194 | No | ||
| 17 | FES | 1427368_x_at 1452410_a_at | 5969 | 0.505 | -0.0134 | No | ||
| 18 | ZFP28 | 1427587_at 1439503_at 1439504_s_at | 7114 | 0.374 | -0.0612 | No | ||
| 19 | HOXD11 | 1450584_at | 7433 | 0.341 | -0.0715 | No | ||
| 20 | CNTN6 | 1422649_at | 7707 | 0.313 | -0.0802 | No | ||
| 21 | SF3A1 | 1419087_s_at 1445719_at 1446687_at 1449333_at | 7965 | 0.290 | -0.0884 | No | ||
| 22 | AKT1S1 | 1428158_at 1437736_at 1452684_at | 7976 | 0.289 | -0.0853 | No | ||
| 23 | PCSK2 | 1428305_at 1444147_at 1447991_at 1447992_s_at 1448312_at | 8182 | 0.268 | -0.0914 | No | ||
| 24 | RDH12 | 1424256_at 1431010_a_at | 8248 | 0.261 | -0.0912 | No | ||
| 25 | MTR | 1439811_at 1458532_at | 8321 | 0.254 | -0.0914 | No | ||
| 26 | DPYSL5 | 1441246_s_at 1449290_at | 8896 | 0.200 | -0.1152 | No | ||
| 27 | SYT3 | 1417708_at | 8971 | 0.192 | -0.1162 | No | ||
| 28 | CFL1 | 1448346_at 1455138_x_at | 9251 | 0.167 | -0.1270 | No | ||
| 29 | GRK5 | 1446361_at 1446998_at 1449514_at | 9571 | 0.139 | -0.1399 | No | ||
| 30 | ABLIM2 | 1435441_at 1455313_at | 10000 | 0.101 | -0.1582 | No | ||
| 31 | TLE4 | 1425650_at 1430384_at 1450853_at | 10111 | 0.093 | -0.1621 | No | ||
| 32 | TSNAXIP1 | 1427018_at | 11100 | 0.008 | -0.2072 | No | ||
| 33 | STMN1 | 1415849_s_at | 11332 | -0.013 | -0.2176 | No | ||
| 34 | ABCC5 | 1418042_a_at 1418043_at 1427565_a_at 1435683_a_at 1435684_at 1435685_x_at 1438056_x_at 1447384_at 1459782_x_at | 11341 | -0.014 | -0.2178 | No | ||
| 35 | ZFP1 | 1427702_at | 11490 | -0.027 | -0.2243 | No | ||
| 36 | SIX1 | 1427277_at | 11722 | -0.048 | -0.2343 | No | ||
| 37 | HSD3B7 | 1416968_a_at 1442912_at | 11827 | -0.058 | -0.2383 | No | ||
| 38 | UBE2N | 1422559_at 1435384_at | 11860 | -0.060 | -0.2390 | No | ||
| 39 | USP48 | 1419277_at 1419278_at 1424056_at | 12155 | -0.085 | -0.2514 | No | ||
| 40 | CACNA1G | 1423365_at | 12328 | -0.101 | -0.2581 | No | ||
| 41 | NCDN | 1416852_a_at 1416853_at | 12697 | -0.134 | -0.2733 | No | ||
| 42 | MAX | 1423501_at | 12735 | -0.138 | -0.2733 | No | ||
| 43 | PPP2R5B | 1426811_at | 13247 | -0.186 | -0.2944 | No | ||
| 44 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 13272 | -0.188 | -0.2932 | No | ||
| 45 | WHSC1L1 | 1442085_at 1447931_at 1455668_at 1457630_at 1457793_a_at 1457794_at 1459907_a_at | 13780 | -0.233 | -0.3135 | No | ||
| 46 | RASSF2 | 1428392_at 1444889_at | 14289 | -0.285 | -0.3333 | No | ||
| 47 | ACBD5 | 1428236_at 1446433_at | 14759 | -0.336 | -0.3506 | No | ||
| 48 | GSK3A | 1435638_at | 14970 | -0.359 | -0.3558 | No | ||
| 49 | NUTF2 | 1434367_s_at 1447440_at 1449017_at | 14990 | -0.361 | -0.3523 | No | ||
| 50 | BAT4 | 1420284_at 1423163_at | 15739 | -0.447 | -0.3810 | No | ||
| 51 | SP2 | 1426237_at 1447730_at | 15810 | -0.455 | -0.3787 | No | ||
| 52 | EBF2 | 1418494_at 1446128_at 1449101_at 1449102_at | 16180 | -0.502 | -0.3894 | No | ||
| 53 | SLC16A4 | 1426082_a_at | 16222 | -0.507 | -0.3851 | No | ||
| 54 | BLCAP | 1420631_a_at | 16382 | -0.528 | -0.3859 | No | ||
| 55 | FYCO1 | 1427177_at 1453424_at | 16544 | -0.552 | -0.3865 | No | ||
| 56 | ARL6IP2 | 1416793_at 1416794_at 1431197_at 1435594_at | 17076 | -0.632 | -0.4031 | No | ||
| 57 | PAK3 | 1417923_at 1417924_at 1437318_at | 17594 | -0.720 | -0.4179 | No | ||
| 58 | MRPL24 | 1428709_a_at | 17719 | -0.742 | -0.4145 | No | ||
| 59 | DAZAP1 | 1419275_at | 17938 | -0.788 | -0.4148 | No | ||
| 60 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 18863 | -0.995 | -0.4449 | Yes | ||
| 61 | NTAN1 | 1423538_at 1447917_x_at | 18875 | -0.998 | -0.4332 | Yes | ||
| 62 | ASXL1 | 1434561_at 1435077_at 1458380_at | 18985 | -1.014 | -0.4258 | Yes | ||
| 63 | TBC1D17 | 1423659_a_at | 19126 | -1.037 | -0.4195 | Yes | ||
| 64 | TIAL1 | 1421148_a_at 1426352_s_at 1446713_at 1452821_at 1455675_a_at 1455676_x_at | 19273 | -1.071 | -0.4131 | Yes | ||
| 65 | IRF2BP1 | 1451252_at | 19692 | -1.195 | -0.4176 | Yes | ||
| 66 | PHF13 | 1455175_at | 19835 | -1.246 | -0.4089 | Yes | ||
| 67 | FEN1 | 1421731_a_at 1436454_x_at | 19915 | -1.276 | -0.3968 | Yes | ||
| 68 | DPF1 | 1420529_at 1420530_at 1442440_at | 20270 | -1.446 | -0.3953 | Yes | ||
| 69 | IPO7 | 1442511_at 1454955_at 1458165_at | 20325 | -1.476 | -0.3797 | Yes | ||
| 70 | CLSPN | 1441984_at 1456280_at | 20474 | -1.547 | -0.3676 | Yes | ||
| 71 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 20490 | -1.553 | -0.3493 | Yes | ||
| 72 | SDCCAG8 | 1431203_at 1444831_at 1453946_a_at 1456538_at | 20583 | -1.615 | -0.3337 | Yes | ||
| 73 | TRUB1 | 1428281_at 1432019_at | 20710 | -1.701 | -0.3186 | Yes | ||
| 74 | RANBP10 | 1424584_a_at 1424585_at 1446446_at | 20847 | -1.794 | -0.3029 | Yes | ||
| 75 | SOLH | 1427785_x_at 1434416_a_at 1439306_at 1450817_at | 21151 | -2.028 | -0.2920 | Yes | ||
| 76 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 21199 | -2.056 | -0.2690 | Yes | ||
| 77 | TTLL4 | 1428911_at 1428912_at 1436249_at 1459031_at | 21271 | -2.145 | -0.2460 | Yes | ||
| 78 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 21275 | -2.151 | -0.2198 | Yes | ||
| 79 | SYMPK | 1428673_at | 21485 | -2.443 | -0.1994 | Yes | ||
| 80 | MADD | 1425735_at 1455502_at | 21698 | -2.944 | -0.1731 | Yes | ||
| 81 | GTPBP2 | 1416690_at 1416691_at 1442305_at 1448437_a_at | 21914 | -7.408 | -0.0923 | Yes | ||
| 82 | FNTB | 1434309_at 1459043_at | 21917 | -7.619 | 0.0009 | Yes |

