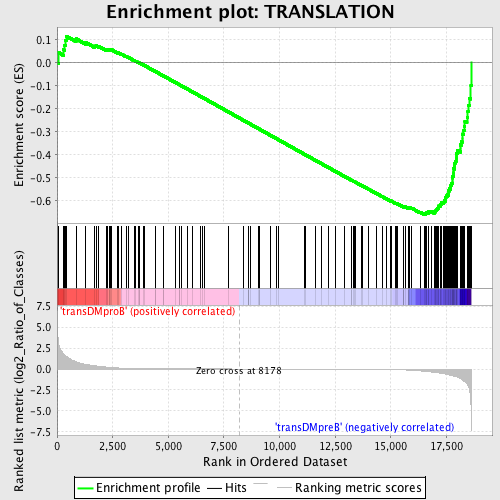
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | TRANSLATION |
| Enrichment Score (ES) | -0.6587271 |
| Normalized Enrichment Score (NES) | -1.5801795 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1899315 |
| FWER p-Value | 0.676 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL3L | 23354 | 51 | 3.466 | 0.0451 | No | ||
| 2 | EBI3 | 23193 | 288 | 1.779 | 0.0569 | No | ||
| 3 | HBS1L | 20076 12127 7119 | 343 | 1.636 | 0.0766 | No | ||
| 4 | APBB1 | 17705 | 373 | 1.578 | 0.0969 | No | ||
| 5 | LTB | 23259 | 410 | 1.533 | 0.1161 | No | ||
| 6 | AARS | 10630 6151 | 853 | 0.873 | 0.1042 | No | ||
| 7 | SPN | 452 5493 | 1286 | 0.550 | 0.0885 | No | ||
| 8 | PAIP2 | 12593 | 1668 | 0.389 | 0.0732 | No | ||
| 9 | EIF2AK3 | 17421 | 1750 | 0.362 | 0.0738 | No | ||
| 10 | EEF2K | 18101 | 1858 | 0.324 | 0.0725 | No | ||
| 11 | BCL10 | 15397 | 2237 | 0.220 | 0.0551 | No | ||
| 12 | NDUFA13 | 7400 | 2248 | 0.217 | 0.0576 | No | ||
| 13 | TLR7 | 24004 | 2341 | 0.193 | 0.0552 | No | ||
| 14 | RPL28 | 5392 | 2376 | 0.185 | 0.0560 | No | ||
| 15 | RPL22 | 15979 | 2398 | 0.181 | 0.0573 | No | ||
| 16 | RPL11 | 12450 | 2440 | 0.173 | 0.0575 | No | ||
| 17 | RPL15 | 12355 | 2730 | 0.125 | 0.0436 | No | ||
| 18 | EIF2B2 | 21204 | 2775 | 0.116 | 0.0428 | No | ||
| 19 | RPL19 | 9736 | 2905 | 0.100 | 0.0372 | No | ||
| 20 | RRBP1 | 8176 14411 | 3107 | 0.081 | 0.0274 | No | ||
| 21 | FURIN | 9538 | 3189 | 0.072 | 0.0241 | No | ||
| 22 | RPL7 | 9748 | 3458 | 0.052 | 0.0103 | No | ||
| 23 | TLR6 | 215 16528 | 3532 | 0.048 | 0.0070 | No | ||
| 24 | RPL39 | 12496 | 3657 | 0.042 | 0.0009 | No | ||
| 25 | GHRL | 1183 17040 | 3710 | 0.039 | -0.0014 | No | ||
| 26 | EIF4H | 16352 | 3718 | 0.039 | -0.0013 | No | ||
| 27 | TNIP1 | 12192 20450 1320 | 3860 | 0.034 | -0.0084 | No | ||
| 28 | RPS11 | 11317 | 3922 | 0.031 | -0.0113 | No | ||
| 29 | EIF2C1 | 10672 | 4422 | 0.018 | -0.0380 | No | ||
| 30 | RPL13 | 11284 | 4430 | 0.018 | -0.0382 | No | ||
| 31 | CD28 | 14239 4092 | 4769 | 0.014 | -0.0563 | No | ||
| 32 | RPL5 | 9746 | 4800 | 0.014 | -0.0577 | No | ||
| 33 | RPS10 | 12464 | 5333 | 0.010 | -0.0864 | No | ||
| 34 | TLR3 | 18884 | 5492 | 0.009 | -0.0948 | No | ||
| 35 | DDX25 | 19184 | 5505 | 0.009 | -0.0953 | No | ||
| 36 | TSC1 | 7235 | 5583 | 0.008 | -0.0994 | No | ||
| 37 | INHA | 14214 | 5844 | 0.007 | -0.1134 | No | ||
| 38 | RPL35 | 12360 | 6071 | 0.006 | -0.1255 | No | ||
| 39 | SAMD4A | 13211 | 6450 | 0.005 | -0.1459 | No | ||
| 40 | INHBB | 13858 | 6549 | 0.004 | -0.1512 | No | ||
| 41 | RPS27 | 12183 | 6613 | 0.004 | -0.1545 | No | ||
| 42 | TNFRSF8 | 2373 5784 | 7712 | 0.001 | -0.2139 | No | ||
| 43 | BACE2 | 1738 22687 1695 | 8391 | -0.001 | -0.2506 | No | ||
| 44 | IL12B | 20918 | 8591 | -0.001 | -0.2614 | No | ||
| 45 | IL27 | 17636 | 8592 | -0.001 | -0.2614 | No | ||
| 46 | RPS3 | 6549 11295 | 8616 | -0.001 | -0.2626 | No | ||
| 47 | DAZL | 22944 | 8672 | -0.001 | -0.2656 | No | ||
| 48 | IL9 | 21444 | 9030 | -0.002 | -0.2848 | No | ||
| 49 | EIF4A2 | 4660 1679 1645 | 9078 | -0.002 | -0.2874 | No | ||
| 50 | MTRF1 | 3413 21949 | 9097 | -0.002 | -0.2883 | No | ||
| 51 | GLMN | 16452 3562 | 9611 | -0.003 | -0.3160 | No | ||
| 52 | TLR9 | 19331 3127 | 9865 | -0.004 | -0.3297 | No | ||
| 53 | IL17F | 13992 | 9959 | -0.004 | -0.3346 | No | ||
| 54 | EREG | 4679 16797 | 11098 | -0.007 | -0.3961 | No | ||
| 55 | TLR8 | 9308 | 11176 | -0.008 | -0.4002 | No | ||
| 56 | RPS5 | 18391 | 11625 | -0.009 | -0.4243 | No | ||
| 57 | YBX2 | 20818 | 11868 | -0.011 | -0.4373 | No | ||
| 58 | EIF2AK4 | 14909 | 12182 | -0.012 | -0.4540 | No | ||
| 59 | YARS | 16071 | 12527 | -0.014 | -0.4725 | No | ||
| 60 | RPL26 | 9739 | 12927 | -0.017 | -0.4938 | No | ||
| 61 | IL6 | 16895 | 13211 | -0.019 | -0.5089 | No | ||
| 62 | RPS6KB2 | 7198 | 13235 | -0.020 | -0.5099 | No | ||
| 63 | SFTPD | 21867 | 13329 | -0.020 | -0.5146 | No | ||
| 64 | RPL18 | 450 5390 | 13376 | -0.021 | -0.5168 | No | ||
| 65 | IL4 | 9174 | 13402 | -0.021 | -0.5179 | No | ||
| 66 | ELA2 | 11925 8893 | 13684 | -0.025 | -0.5327 | No | ||
| 67 | BOLL | 7960 | 13722 | -0.026 | -0.5344 | No | ||
| 68 | TLR1 | 10190 | 13990 | -0.031 | -0.5484 | No | ||
| 69 | GHSR | 1810 15625 | 14369 | -0.038 | -0.5683 | No | ||
| 70 | RPL41 | 12611 | 14629 | -0.046 | -0.5817 | No | ||
| 71 | TLR4 | 2329 10191 5770 | 14788 | -0.051 | -0.5896 | No | ||
| 72 | RPL37 | 12502 7421 22521 | 14998 | -0.061 | -0.6000 | No | ||
| 73 | FOXP3 | 9804 5426 | 15013 | -0.062 | -0.5999 | No | ||
| 74 | MAST2 | 9423 | 15200 | -0.073 | -0.6090 | No | ||
| 75 | PAIP1 | 21556 | 15276 | -0.078 | -0.6120 | No | ||
| 76 | RPL24 | 12664 | 15317 | -0.080 | -0.6131 | No | ||
| 77 | HSPB1 | 4879 | 15573 | -0.101 | -0.6255 | No | ||
| 78 | RPS17 | 9753 | 15582 | -0.102 | -0.6245 | No | ||
| 79 | RPL3 | 11330 | 15584 | -0.102 | -0.6231 | No | ||
| 80 | FARS2 | 21666 | 15647 | -0.110 | -0.6250 | No | ||
| 81 | APOA2 | 8615 4044 | 15777 | -0.125 | -0.6302 | No | ||
| 82 | EIF4G3 | 10517 | 15792 | -0.127 | -0.6292 | No | ||
| 83 | MTIF2 | 13354 | 15813 | -0.130 | -0.6285 | No | ||
| 84 | RPL8 | 22437 | 15856 | -0.136 | -0.6289 | No | ||
| 85 | RPL18A | 13358 | 15931 | -0.146 | -0.6309 | No | ||
| 86 | RPL23A | 11193 | 16315 | -0.221 | -0.6486 | No | ||
| 87 | BCL3 | 8654 | 16492 | -0.266 | -0.6544 | No | ||
| 88 | SARS2 | 5 | 16573 | -0.287 | -0.6548 | Yes | ||
| 89 | RPL7A | 11312 | 16603 | -0.294 | -0.6523 | Yes | ||
| 90 | EEF1A1 | 38 38 8879 | 16614 | -0.296 | -0.6487 | Yes | ||
| 91 | RPS3A | 9755 | 16692 | -0.318 | -0.6485 | Yes | ||
| 92 | MRPL51 | 12361 7301 | 16699 | -0.319 | -0.6444 | Yes | ||
| 93 | EIF4G2 | 1908 8892 | 16813 | -0.350 | -0.6457 | Yes | ||
| 94 | COPS5 | 4117 14002 | 16947 | -0.390 | -0.6475 | Yes | ||
| 95 | EIF4A3 | 9647 | 16988 | -0.404 | -0.6441 | Yes | ||
| 96 | EIF4B | 13279 7979 | 17024 | -0.419 | -0.6402 | Yes | ||
| 97 | RPL6 | 9747 | 17067 | -0.434 | -0.6365 | Yes | ||
| 98 | RPS19 | 5398 | 17101 | -0.445 | -0.6321 | Yes | ||
| 99 | RPS2 | 9279 | 17144 | -0.459 | -0.6280 | Yes | ||
| 100 | NACA | 9444 5147 | 17147 | -0.459 | -0.6218 | Yes | ||
| 101 | MRPL23 | 9738 | 17217 | -0.486 | -0.6188 | Yes | ||
| 102 | MRPL10 | 20681 | 17245 | -0.503 | -0.6133 | Yes | ||
| 103 | METAP1 | 15154 | 17267 | -0.511 | -0.6074 | Yes | ||
| 104 | RPL30 | 9742 | 17366 | -0.561 | -0.6049 | Yes | ||
| 105 | SCYE1 | 8897 | 17394 | -0.575 | -0.5985 | Yes | ||
| 106 | PET112L | 1824 15563 1819 | 17445 | -0.594 | -0.5930 | Yes | ||
| 107 | MRPL55 | 20864 | 17456 | -0.602 | -0.5852 | Yes | ||
| 108 | RPL31 | 14267 | 17485 | -0.621 | -0.5781 | Yes | ||
| 109 | IL10 | 14145 1510 1553 22902 | 17539 | -0.650 | -0.5720 | Yes | ||
| 110 | MRPS7 | 20595 | 17599 | -0.689 | -0.5657 | Yes | ||
| 111 | MRPL52 | 22019 | 17601 | -0.691 | -0.5562 | Yes | ||
| 112 | IRF4 | 21679 | 17618 | -0.695 | -0.5474 | Yes | ||
| 113 | UPF1 | 9718 18855 | 17675 | -0.729 | -0.5404 | Yes | ||
| 114 | EIF2AK1 | 4869 | 17694 | -0.738 | -0.5312 | Yes | ||
| 115 | DHPS | 3815 | 17728 | -0.760 | -0.5225 | Yes | ||
| 116 | MRPS24 | 12262 | 17776 | -0.799 | -0.5140 | Yes | ||
| 117 | KARS | 18738 | 17777 | -0.800 | -0.5029 | Yes | ||
| 118 | ABCF1 | 22996 10329 5897 | 17782 | -0.803 | -0.4920 | Yes | ||
| 119 | RPL14 | 19267 | 17812 | -0.822 | -0.4822 | Yes | ||
| 120 | DPH1 | 4375 8580 8581 | 17821 | -0.831 | -0.4712 | Yes | ||
| 121 | EIF2B4 | 16574 | 17823 | -0.831 | -0.4598 | Yes | ||
| 122 | EIF2B5 | 1719 22822 | 17856 | -0.859 | -0.4496 | Yes | ||
| 123 | TUFM | 10608 | 17869 | -0.868 | -0.4383 | Yes | ||
| 124 | WARS | 2085 20984 | 17890 | -0.885 | -0.4271 | Yes | ||
| 125 | RPL21 | 9737 16624 5391 | 17936 | -0.918 | -0.4169 | Yes | ||
| 126 | RPL29 | 9741 | 17947 | -0.928 | -0.4046 | Yes | ||
| 127 | ETF1 | 23467 | 17951 | -0.929 | -0.3919 | Yes | ||
| 128 | PRG3 | 14971 | 18005 | -0.984 | -0.3812 | Yes | ||
| 129 | RPL4 | 7499 19411 | 18126 | -1.136 | -0.3720 | Yes | ||
| 130 | FARSB | 13909 | 18127 | -1.136 | -0.3563 | Yes | ||
| 131 | EIF2B1 | 16368 3458 | 18163 | -1.187 | -0.3418 | Yes | ||
| 132 | EIF1 | 23563 | 18209 | -1.270 | -0.3266 | Yes | ||
| 133 | RARS | 20496 | 18210 | -1.272 | -0.3091 | Yes | ||
| 134 | HRSP12 | 9123 9122 4870 9638 | 18279 | -1.407 | -0.2933 | Yes | ||
| 135 | EIF5A | 11345 20379 6590 | 18300 | -1.448 | -0.2744 | Yes | ||
| 136 | DARS | 10375 13846 | 18330 | -1.506 | -0.2551 | Yes | ||
| 137 | EIF2B3 | 16118 | 18425 | -1.775 | -0.2357 | Yes | ||
| 138 | RPL27A | 11181 6467 18130 | 18462 | -1.936 | -0.2109 | Yes | ||
| 139 | EIF5 | 5736 | 18479 | -2.027 | -0.1837 | Yes | ||
| 140 | MRPL3 | 3116 19335 | 18528 | -2.380 | -0.1535 | Yes | ||
| 141 | PABPC4 | 10510 2418 2548 | 18600 | -4.171 | -0.0997 | Yes | ||
| 142 | RPL38 | 12562 20606 7475 | 18616 | -7.271 | 0.0000 | Yes |

