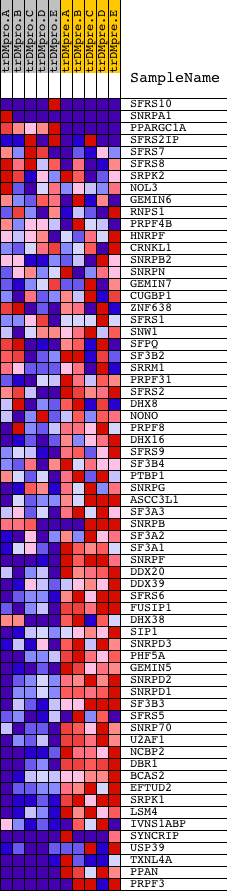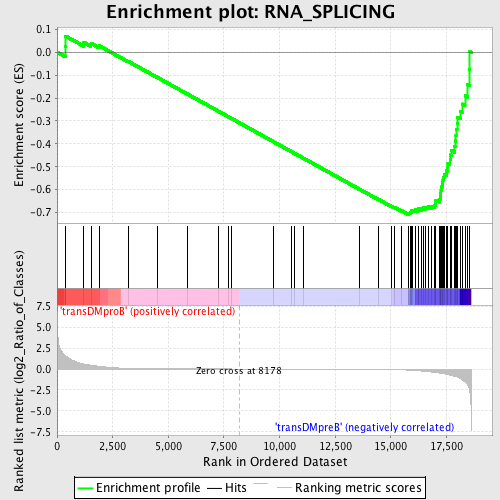
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | RNA_SPLICING |
| Enrichment Score (ES) | -0.7081641 |
| Normalized Enrichment Score (NES) | -1.5480729 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.20288548 |
| FWER p-Value | 0.878 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 9818 1722 5441 | 368 | 1.583 | 0.0257 | No | ||
| 2 | SNRPA1 | 1373 18216 | 381 | 1.570 | 0.0702 | No | ||
| 3 | PPARGC1A | 16533 | 1193 | 0.597 | 0.0437 | No | ||
| 4 | SFRS2IP | 7794 13009 | 1523 | 0.451 | 0.0389 | No | ||
| 5 | SFRS7 | 22889 | 1898 | 0.314 | 0.0278 | No | ||
| 6 | SFRS8 | 10543 6089 | 3195 | 0.072 | -0.0400 | No | ||
| 7 | SRPK2 | 5513 | 4494 | 0.017 | -0.1095 | No | ||
| 8 | NOL3 | 18496 915 921 | 5882 | 0.007 | -0.1840 | No | ||
| 9 | GEMIN6 | 12492 7415 | 7238 | 0.002 | -0.2570 | No | ||
| 10 | RNPS1 | 9730 23361 | 7696 | 0.001 | -0.2816 | No | ||
| 11 | PRPF4B | 3208 5295 | 7854 | 0.001 | -0.2901 | No | ||
| 12 | HNRPF | 13607 | 9741 | -0.004 | -0.3916 | No | ||
| 13 | CRNKL1 | 14407 | 10515 | -0.006 | -0.4331 | No | ||
| 14 | SNRPB2 | 5470 | 10690 | -0.006 | -0.4423 | No | ||
| 15 | SNRPN | 1715 9844 5471 | 11086 | -0.007 | -0.4634 | No | ||
| 16 | GEMIN7 | 17944 | 13580 | -0.024 | -0.5971 | No | ||
| 17 | CUGBP1 | 2805 8819 4576 2924 | 14465 | -0.041 | -0.6436 | No | ||
| 18 | ZNF638 | 9476 | 15023 | -0.063 | -0.6718 | No | ||
| 19 | SFRS1 | 8492 | 15175 | -0.071 | -0.6779 | No | ||
| 20 | SNW1 | 7282 | 15501 | -0.094 | -0.6927 | No | ||
| 21 | SFPQ | 12936 | 15789 | -0.127 | -0.7045 | Yes | ||
| 22 | SF3B2 | 23974 | 15809 | -0.129 | -0.7018 | Yes | ||
| 23 | SRRM1 | 6958 | 15872 | -0.139 | -0.7012 | Yes | ||
| 24 | PRPF31 | 7594 | 15904 | -0.143 | -0.6987 | Yes | ||
| 25 | SFRS2 | 9807 20136 | 15923 | -0.146 | -0.6955 | Yes | ||
| 26 | DHX8 | 20649 | 15940 | -0.148 | -0.6921 | Yes | ||
| 27 | NONO | 11993 | 15961 | -0.152 | -0.6888 | Yes | ||
| 28 | PRPF8 | 20780 1371 | 16105 | -0.178 | -0.6914 | Yes | ||
| 29 | DHX16 | 23247 | 16112 | -0.179 | -0.6866 | Yes | ||
| 30 | SFRS9 | 16731 | 16235 | -0.203 | -0.6873 | Yes | ||
| 31 | SF3B4 | 22269 | 16255 | -0.207 | -0.6824 | Yes | ||
| 32 | PTBP1 | 5303 | 16386 | -0.238 | -0.6825 | Yes | ||
| 33 | SNRPG | 12622 | 16463 | -0.258 | -0.6792 | Yes | ||
| 34 | ASCC3L1 | 14867 | 16562 | -0.285 | -0.6763 | Yes | ||
| 35 | SF3A3 | 16091 | 16675 | -0.313 | -0.6733 | Yes | ||
| 36 | SNRPB | 9842 5469 2736 | 16845 | -0.360 | -0.6721 | Yes | ||
| 37 | SF3A2 | 19938 | 16967 | -0.396 | -0.6672 | Yes | ||
| 38 | SF3A1 | 7450 | 17019 | -0.417 | -0.6579 | Yes | ||
| 39 | SNRPF | 7645 | 17025 | -0.419 | -0.6461 | Yes | ||
| 40 | DDX20 | 15213 | 17173 | -0.466 | -0.6406 | Yes | ||
| 41 | DDX39 | 18551 | 17216 | -0.486 | -0.6289 | Yes | ||
| 42 | SFRS6 | 14751 | 17246 | -0.503 | -0.6160 | Yes | ||
| 43 | FUSIP1 | 4715 16036 | 17254 | -0.507 | -0.6018 | Yes | ||
| 44 | DHX38 | 48 | 17271 | -0.512 | -0.5879 | Yes | ||
| 45 | SIP1 | 21263 | 17307 | -0.533 | -0.5745 | Yes | ||
| 46 | SNRPD3 | 12514 | 17340 | -0.547 | -0.5605 | Yes | ||
| 47 | PHF5A | 22194 | 17362 | -0.559 | -0.5455 | Yes | ||
| 48 | GEMIN5 | 20439 | 17423 | -0.586 | -0.5319 | Yes | ||
| 49 | SNRPD2 | 8412 | 17505 | -0.632 | -0.5181 | Yes | ||
| 50 | SNRPD1 | 23622 | 17533 | -0.647 | -0.5009 | Yes | ||
| 51 | SF3B3 | 18746 | 17567 | -0.669 | -0.4835 | Yes | ||
| 52 | SFRS5 | 9808 2062 | 17659 | -0.716 | -0.4678 | Yes | ||
| 53 | SNRP70 | 1186 | 17695 | -0.739 | -0.4484 | Yes | ||
| 54 | U2AF1 | 8431 | 17718 | -0.750 | -0.4280 | Yes | ||
| 55 | NCBP2 | 12643 | 17873 | -0.871 | -0.4113 | Yes | ||
| 56 | DBR1 | 19341 | 17894 | -0.890 | -0.3867 | Yes | ||
| 57 | BCAS2 | 12662 | 17926 | -0.912 | -0.3622 | Yes | ||
| 58 | EFTUD2 | 1219 20203 | 17956 | -0.931 | -0.3370 | Yes | ||
| 59 | SRPK1 | 23049 | 17977 | -0.952 | -0.3106 | Yes | ||
| 60 | LSM4 | 18583 | 17992 | -0.964 | -0.2836 | Yes | ||
| 61 | IVNS1ABP | 4384 4050 | 18124 | -1.135 | -0.2580 | Yes | ||
| 62 | SYNCRIP | 3078 3035 3107 | 18203 | -1.259 | -0.2260 | Yes | ||
| 63 | USP39 | 1116 1083 11373 | 18335 | -1.516 | -0.1894 | Yes | ||
| 64 | TXNL4A | 6567 11329 | 18451 | -1.865 | -0.1420 | Yes | ||
| 65 | PPAN | 19546 | 18541 | -2.549 | -0.0734 | Yes | ||
| 66 | PRPF3 | 12899 12898 1877 7703 | 18551 | -2.691 | 0.0035 | Yes |