Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
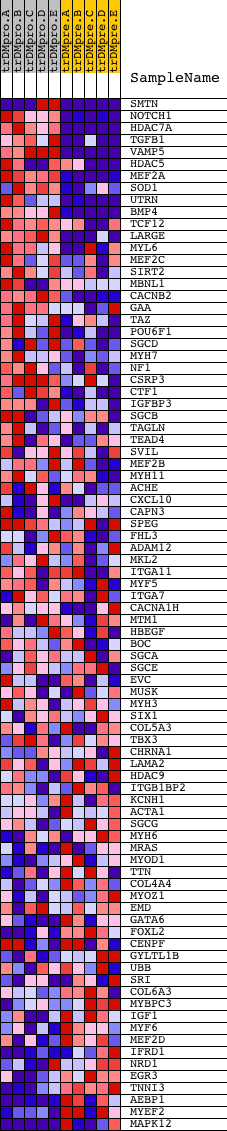
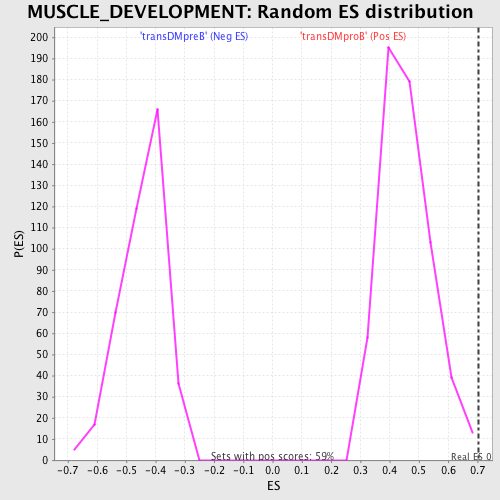
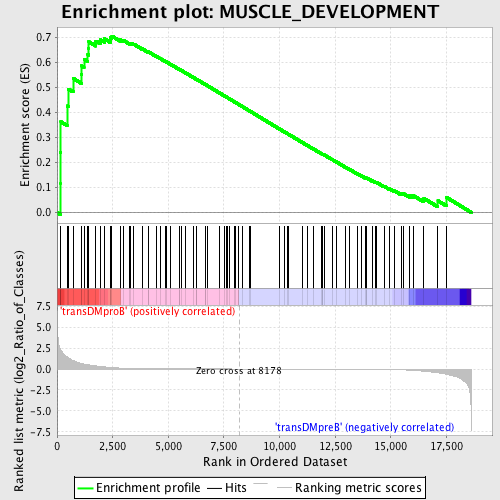
| GeneSet | MUSCLE_DEVELOPMENT |
| Enrichment Score (ES) | 0.70485085 |
| Normalized Enrichment Score (NES) | 1.5416615 |
| Nominal p-value | 0.0017035775 |
| FDR q-value | 0.29158446 |
| FWER p-Value | 0.872 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMTN | 20551 1446 | 152 | 2.313 | 0.1171 | Yes | ||
| 2 | NOTCH1 | 14649 | 158 | 2.287 | 0.2408 | Yes | ||
| 3 | HDAC7A | 22147 | 170 | 2.250 | 0.3621 | Yes | ||
| 4 | TGFB1 | 18332 | 452 | 1.452 | 0.4257 | Yes | ||
| 5 | VAMP5 | 17118 | 515 | 1.305 | 0.4930 | Yes | ||
| 6 | HDAC5 | 1480 20205 | 723 | 0.996 | 0.5358 | Yes | ||
| 7 | MEF2A | 1201 5089 3492 | 1095 | 0.656 | 0.5514 | Yes | ||
| 8 | SOD1 | 9846 | 1109 | 0.648 | 0.5858 | Yes | ||
| 9 | UTRN | 5835 | 1211 | 0.588 | 0.6122 | Yes | ||
| 10 | BMP4 | 21863 | 1381 | 0.510 | 0.6308 | Yes | ||
| 11 | TCF12 | 10044 3125 | 1417 | 0.495 | 0.6557 | Yes | ||
| 12 | LARGE | 18837 | 1426 | 0.492 | 0.6819 | Yes | ||
| 13 | MYL6 | 9438 3408 | 1737 | 0.366 | 0.6850 | Yes | ||
| 14 | MEF2C | 3204 9378 | 1945 | 0.302 | 0.6902 | Yes | ||
| 15 | SIRT2 | 18318 | 2125 | 0.249 | 0.6941 | Yes | ||
| 16 | MBNL1 | 1921 15582 | 2390 | 0.183 | 0.6897 | Yes | ||
| 17 | CACNB2 | 4466 8678 | 2406 | 0.180 | 0.6987 | Yes | ||
| 18 | GAA | 20575 | 2462 | 0.169 | 0.7049 | Yes | ||
| 19 | TAZ | 12415 | 2847 | 0.107 | 0.6899 | No | ||
| 20 | POU6F1 | 22128 | 2979 | 0.092 | 0.6879 | No | ||
| 21 | SGCD | 1425 10779 | 3269 | 0.066 | 0.6758 | No | ||
| 22 | MYH7 | 21828 4709 | 3305 | 0.063 | 0.6773 | No | ||
| 23 | NF1 | 5165 | 3443 | 0.054 | 0.6728 | No | ||
| 24 | CSRP3 | 17813 1261 5107 16068 5106 9394 | 3819 | 0.035 | 0.6545 | No | ||
| 25 | CTF1 | 18064 | 4095 | 0.026 | 0.6411 | No | ||
| 26 | IGFBP3 | 4898 | 4100 | 0.026 | 0.6423 | No | ||
| 27 | SGCB | 10777 6276 | 4483 | 0.017 | 0.6226 | No | ||
| 28 | TAGLN | 19136 | 4645 | 0.015 | 0.6148 | No | ||
| 29 | TEAD4 | 997 5708 | 4850 | 0.013 | 0.6045 | No | ||
| 30 | SVIL | 1990 1942 10334 1976 23626 1946 1945 2041 | 4938 | 0.012 | 0.6004 | No | ||
| 31 | MEF2B | 9377 | 5115 | 0.011 | 0.5915 | No | ||
| 32 | MYH11 | 22656 | 5488 | 0.009 | 0.5719 | No | ||
| 33 | ACHE | 16665 | 5511 | 0.009 | 0.5712 | No | ||
| 34 | CXCL10 | 16477 | 5591 | 0.008 | 0.5674 | No | ||
| 35 | CAPN3 | 8686 2868 | 5780 | 0.007 | 0.5576 | No | ||
| 36 | SPEG | 8608 | 6141 | 0.006 | 0.5385 | No | ||
| 37 | FHL3 | 16092 2457 | 6248 | 0.005 | 0.5331 | No | ||
| 38 | ADAM12 | 2207 8545 | 6278 | 0.005 | 0.5318 | No | ||
| 39 | MKL2 | 10746 | 6668 | 0.004 | 0.5110 | No | ||
| 40 | ITGA11 | 6677 11449 | 6669 | 0.004 | 0.5112 | No | ||
| 41 | MYF5 | 19632 | 6749 | 0.004 | 0.5072 | No | ||
| 42 | ITGA7 | 19835 | 7306 | 0.002 | 0.4773 | No | ||
| 43 | CACNA1H | 23077 | 7540 | 0.002 | 0.4648 | No | ||
| 44 | MTM1 | 9422 2634 | 7625 | 0.001 | 0.4603 | No | ||
| 45 | HBEGF | 23457 | 7654 | 0.001 | 0.4589 | No | ||
| 46 | BOC | 22587 | 7741 | 0.001 | 0.4543 | No | ||
| 47 | SGCA | 20286 1216 | 7984 | 0.000 | 0.4413 | No | ||
| 48 | SGCE | 17225 | 8036 | 0.000 | 0.4386 | No | ||
| 49 | EVC | 7206 | 8133 | 0.000 | 0.4334 | No | ||
| 50 | MUSK | 5199 | 8155 | 0.000 | 0.4323 | No | ||
| 51 | MYH3 | 20839 | 8310 | -0.000 | 0.4240 | No | ||
| 52 | SIX1 | 9821 | 8647 | -0.001 | 0.4059 | No | ||
| 53 | COL5A3 | 19219 | 8698 | -0.001 | 0.4033 | No | ||
| 54 | TBX3 | 16723 | 10008 | -0.004 | 0.3329 | No | ||
| 55 | CHRNA1 | 4319 14559 8530 | 10207 | -0.005 | 0.3224 | No | ||
| 56 | LAMA2 | 19800 | 10335 | -0.005 | 0.3158 | No | ||
| 57 | HDAC9 | 21084 2131 | 10394 | -0.005 | 0.3130 | No | ||
| 58 | ITGB1BP2 | 24275 | 11010 | -0.007 | 0.2802 | No | ||
| 59 | KCNH1 | 4074 14008 | 11240 | -0.008 | 0.2683 | No | ||
| 60 | ACTA1 | 18715 | 11541 | -0.009 | 0.2526 | No | ||
| 61 | SGCG | 21798 | 11870 | -0.011 | 0.2355 | No | ||
| 62 | MYH6 | 9436 | 11929 | -0.011 | 0.2329 | No | ||
| 63 | MRAS | 9409 | 11948 | -0.011 | 0.2325 | No | ||
| 64 | MYOD1 | 18237 | 12029 | -0.011 | 0.2288 | No | ||
| 65 | TTN | 14552 2743 14553 | 12371 | -0.013 | 0.2111 | No | ||
| 66 | COL4A4 | 8767 | 12545 | -0.014 | 0.2026 | No | ||
| 67 | MYOZ1 | 21907 | 12972 | -0.017 | 0.1805 | No | ||
| 68 | EMD | 2557 24300 | 13124 | -0.019 | 0.1734 | No | ||
| 69 | GATA6 | 96 | 13511 | -0.023 | 0.1538 | No | ||
| 70 | FOXL2 | 19345 | 13680 | -0.025 | 0.1461 | No | ||
| 71 | CENPF | 13723 | 13853 | -0.028 | 0.1383 | No | ||
| 72 | GYLTL1B | 2969 14520 | 13911 | -0.029 | 0.1368 | No | ||
| 73 | UBB | 10240 | 13920 | -0.029 | 0.1379 | No | ||
| 74 | SRI | 3656 8462 | 14171 | -0.034 | 0.1263 | No | ||
| 75 | COL6A3 | 4546 13885 13884 | 14299 | -0.037 | 0.1214 | No | ||
| 76 | MYBPC3 | 9434 | 14363 | -0.038 | 0.1201 | No | ||
| 77 | IGF1 | 3352 9156 3409 | 14702 | -0.048 | 0.1044 | No | ||
| 78 | MYF6 | 19631 | 14922 | -0.057 | 0.0957 | No | ||
| 79 | MEF2D | 9379 | 15172 | -0.071 | 0.0861 | No | ||
| 80 | IFRD1 | 4897 | 15463 | -0.091 | 0.0754 | No | ||
| 81 | NRD1 | 16140 | 15550 | -0.099 | 0.0761 | No | ||
| 82 | EGR3 | 4656 | 15854 | -0.136 | 0.0671 | No | ||
| 83 | TNNI3 | 89 | 15999 | -0.159 | 0.0680 | No | ||
| 84 | AEBP1 | 20953 | 16485 | -0.265 | 0.0561 | No | ||
| 85 | MYEF2 | 5140 | 17119 | -0.453 | 0.0465 | No | ||
| 86 | MAPK12 | 2182 2215 22164 | 17503 | -0.632 | 0.0601 | No |