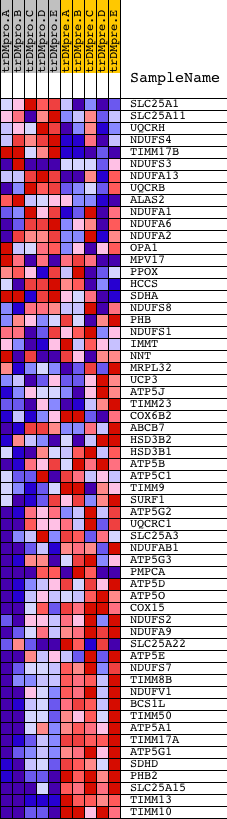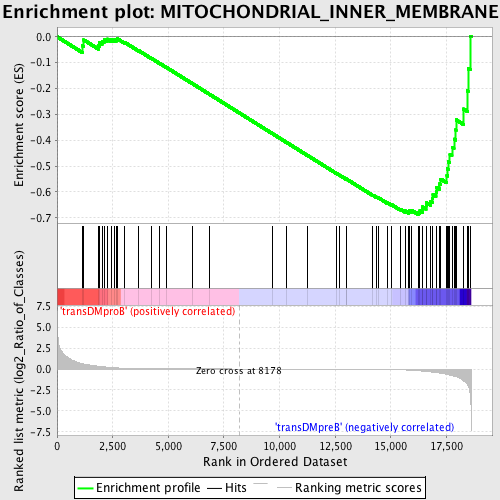
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | -0.6872609 |
| Normalized Enrichment Score (NES) | -1.4986537 |
| Nominal p-value | 0.0022522523 |
| FDR q-value | 0.32687637 |
| FWER p-Value | 0.993 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC25A1 | 22645 | 1148 | 0.619 | -0.0351 | No | ||
| 2 | SLC25A11 | 20366 1368 | 1200 | 0.591 | -0.0122 | No | ||
| 3 | UQCRH | 12378 | 1863 | 0.323 | -0.0339 | No | ||
| 4 | NDUFS4 | 9452 5157 3173 | 1910 | 0.312 | -0.0229 | No | ||
| 5 | TIMM17B | 24390 | 2020 | 0.279 | -0.0167 | No | ||
| 6 | NDUFS3 | 7553 12683 | 2112 | 0.253 | -0.0107 | No | ||
| 7 | NDUFA13 | 7400 | 2248 | 0.217 | -0.0086 | No | ||
| 8 | UQCRB | 12545 | 2443 | 0.173 | -0.0115 | No | ||
| 9 | ALAS2 | 24229 | 2572 | 0.150 | -0.0119 | No | ||
| 10 | NDUFA1 | 24170 | 2659 | 0.136 | -0.0107 | No | ||
| 11 | NDUFA6 | 12473 | 2699 | 0.130 | -0.0072 | No | ||
| 12 | NDUFA2 | 9450 | 3026 | 0.087 | -0.0210 | No | ||
| 13 | OPA1 | 13169 22800 7894 | 3679 | 0.041 | -0.0543 | No | ||
| 14 | MPV17 | 9407 3505 3543 | 4251 | 0.022 | -0.0842 | No | ||
| 15 | PPOX | 13761 4104 | 4603 | 0.016 | -0.1024 | No | ||
| 16 | HCCS | 9079 4842 | 4913 | 0.012 | -0.1185 | No | ||
| 17 | SDHA | 12436 | 6075 | 0.006 | -0.1808 | No | ||
| 18 | NDUFS8 | 23950 | 6853 | 0.003 | -0.2225 | No | ||
| 19 | PHB | 9558 | 9667 | -0.003 | -0.3740 | No | ||
| 20 | NDUFS1 | 13940 | 10312 | -0.005 | -0.4085 | No | ||
| 21 | IMMT | 367 8029 | 11238 | -0.008 | -0.4580 | No | ||
| 22 | NNT | 3273 5181 9471 3238 | 12554 | -0.014 | -0.5282 | No | ||
| 23 | MRPL32 | 7961 | 12710 | -0.015 | -0.5359 | No | ||
| 24 | UCP3 | 3972 18175 | 12989 | -0.017 | -0.5501 | No | ||
| 25 | ATP5J | 880 22555 | 12995 | -0.017 | -0.5496 | No | ||
| 26 | TIMM23 | 7004 5768 | 14181 | -0.034 | -0.6120 | No | ||
| 27 | COX6B2 | 17983 4110 | 14353 | -0.038 | -0.6196 | No | ||
| 28 | ABCB7 | 24085 | 14446 | -0.041 | -0.6228 | No | ||
| 29 | HSD3B2 | 4873 4874 15229 4875 9126 | 14843 | -0.054 | -0.6418 | No | ||
| 30 | HSD3B1 | 15227 | 15042 | -0.064 | -0.6497 | No | ||
| 31 | ATP5B | 19846 | 15422 | -0.088 | -0.6664 | No | ||
| 32 | ATP5C1 | 8635 | 15642 | -0.110 | -0.6734 | No | ||
| 33 | TIMM9 | 19686 | 15812 | -0.130 | -0.6769 | No | ||
| 34 | SURF1 | 14645 | 15825 | -0.132 | -0.6718 | No | ||
| 35 | ATP5G2 | 12610 | 15943 | -0.148 | -0.6717 | No | ||
| 36 | UQCRC1 | 10259 | 16232 | -0.202 | -0.6785 | Yes | ||
| 37 | SLC25A3 | 19644 | 16308 | -0.219 | -0.6731 | Yes | ||
| 38 | NDUFAB1 | 7667 | 16413 | -0.246 | -0.6680 | Yes | ||
| 39 | ATP5G3 | 14558 | 16426 | -0.249 | -0.6579 | Yes | ||
| 40 | PMPCA | 12421 7350 | 16583 | -0.289 | -0.6538 | Yes | ||
| 41 | ATP5D | 19949 | 16598 | -0.292 | -0.6419 | Yes | ||
| 42 | ATP5O | 22539 | 16773 | -0.340 | -0.6366 | Yes | ||
| 43 | COX15 | 5931 | 16880 | -0.370 | -0.6263 | Yes | ||
| 44 | NDUFS2 | 4089 13760 | 16893 | -0.374 | -0.6108 | Yes | ||
| 45 | NDUFA9 | 12288 | 17031 | -0.423 | -0.5998 | Yes | ||
| 46 | SLC25A22 | 12671 | 17059 | -0.431 | -0.5826 | Yes | ||
| 47 | ATP5E | 14321 | 17187 | -0.473 | -0.5690 | Yes | ||
| 48 | NDUFS7 | 19946 | 17242 | -0.502 | -0.5502 | Yes | ||
| 49 | TIMM8B | 11391 | 17521 | -0.640 | -0.5375 | Yes | ||
| 50 | NDUFV1 | 23953 | 17546 | -0.654 | -0.5105 | Yes | ||
| 51 | BCS1L | 14223 | 17581 | -0.677 | -0.4830 | Yes | ||
| 52 | TIMM50 | 17911 | 17657 | -0.716 | -0.4561 | Yes | ||
| 53 | ATP5A1 | 23505 | 17750 | -0.782 | -0.4272 | Yes | ||
| 54 | TIMM17A | 13822 | 17841 | -0.849 | -0.3953 | Yes | ||
| 55 | ATP5G1 | 8636 | 17922 | -0.909 | -0.3603 | Yes | ||
| 56 | SDHD | 19125 | 17934 | -0.918 | -0.3211 | Yes | ||
| 57 | PHB2 | 17286 | 18270 | -1.387 | -0.2792 | Yes | ||
| 58 | SLC25A15 | 356 3819 | 18443 | -1.841 | -0.2088 | Yes | ||
| 59 | TIMM13 | 14974 | 18476 | -2.022 | -0.1230 | Yes | ||
| 60 | TIMM10 | 11390 2158 | 18569 | -3.015 | 0.0025 | Yes |