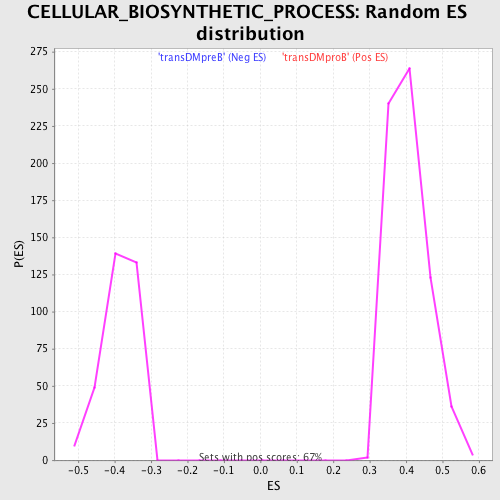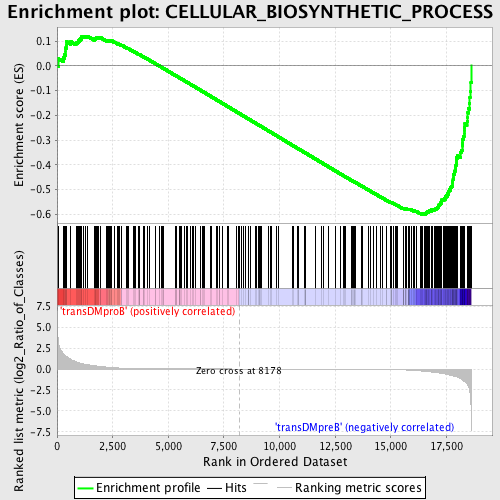
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | CELLULAR_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | -0.60014904 |
| Normalized Enrichment Score (NES) | -1.5508713 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.21607009 |
| FWER p-Value | 0.868 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL3L | 23354 | 51 | 3.466 | 0.0292 | No | ||
| 2 | EBI3 | 23193 | 288 | 1.779 | 0.0327 | No | ||
| 3 | HBS1L | 20076 12127 7119 | 343 | 1.636 | 0.0448 | No | ||
| 4 | FADS1 | 23933 | 369 | 1.583 | 0.0580 | No | ||
| 5 | APBB1 | 17705 | 373 | 1.578 | 0.0724 | No | ||
| 6 | LTB | 23259 | 410 | 1.533 | 0.0846 | No | ||
| 7 | CHST3 | 19760 | 422 | 1.514 | 0.0979 | No | ||
| 8 | DDAH2 | 23263 | 616 | 1.122 | 0.0978 | No | ||
| 9 | AARS | 10630 6151 | 853 | 0.873 | 0.0930 | No | ||
| 10 | TSPO | 2204 22408 | 935 | 0.790 | 0.0958 | No | ||
| 11 | FADS2 | 7127 | 954 | 0.775 | 0.1020 | No | ||
| 12 | EXT1 | 22297 | 997 | 0.734 | 0.1065 | No | ||
| 13 | GMDS | 21496 | 1057 | 0.680 | 0.1095 | No | ||
| 14 | ARPP-19 | 19379 | 1099 | 0.654 | 0.1133 | No | ||
| 15 | SOD1 | 9846 | 1109 | 0.648 | 0.1188 | No | ||
| 16 | PPARGC1A | 16533 | 1193 | 0.597 | 0.1198 | No | ||
| 17 | SPN | 452 5493 | 1286 | 0.550 | 0.1198 | No | ||
| 18 | GCH1 | 9008 4762 | 1366 | 0.517 | 0.1203 | No | ||
| 19 | PAIP2 | 12593 | 1668 | 0.389 | 0.1075 | No | ||
| 20 | AGPAT2 | 2664 | 1720 | 0.372 | 0.1081 | No | ||
| 21 | OAZ1 | 9498 | 1732 | 0.368 | 0.1109 | No | ||
| 22 | EIF2AK3 | 17421 | 1750 | 0.362 | 0.1133 | No | ||
| 23 | HS6ST1 | 14274 | 1812 | 0.339 | 0.1131 | No | ||
| 24 | EEF2K | 18101 | 1858 | 0.324 | 0.1137 | No | ||
| 25 | GAMT | 19698 | 1932 | 0.306 | 0.1125 | No | ||
| 26 | NFE2L1 | 9457 | 1952 | 0.301 | 0.1142 | No | ||
| 27 | BCL10 | 15397 | 2237 | 0.220 | 0.1008 | No | ||
| 28 | NDUFA13 | 7400 | 2248 | 0.217 | 0.1022 | No | ||
| 29 | AP3D1 | 19690 | 2324 | 0.200 | 0.1000 | No | ||
| 30 | TLR7 | 24004 | 2341 | 0.193 | 0.1009 | No | ||
| 31 | RPL28 | 5392 | 2376 | 0.185 | 0.1008 | No | ||
| 32 | RPL22 | 15979 | 2398 | 0.181 | 0.1013 | No | ||
| 33 | HSD3B7 | 4158 | 2436 | 0.174 | 0.1009 | No | ||
| 34 | RPL11 | 12450 | 2440 | 0.173 | 0.1023 | No | ||
| 35 | ALAS2 | 24229 | 2572 | 0.150 | 0.0966 | No | ||
| 36 | RPL15 | 12355 | 2730 | 0.125 | 0.0892 | No | ||
| 37 | EIF2B2 | 21204 | 2775 | 0.116 | 0.0878 | No | ||
| 38 | UROS | 17588 1227 | 2802 | 0.112 | 0.0875 | No | ||
| 39 | DMD | 24295 2647 | 2886 | 0.102 | 0.0839 | No | ||
| 40 | RPL19 | 9736 | 2905 | 0.100 | 0.0838 | No | ||
| 41 | RRBP1 | 8176 14411 | 3107 | 0.081 | 0.0736 | No | ||
| 42 | GLCE | 13526 | 3178 | 0.074 | 0.0705 | No | ||
| 43 | FURIN | 9538 | 3189 | 0.072 | 0.0706 | No | ||
| 44 | PLOD1 | 15679 | 3421 | 0.055 | 0.0585 | No | ||
| 45 | RPL7 | 9748 | 3458 | 0.052 | 0.0570 | No | ||
| 46 | TLR6 | 215 16528 | 3532 | 0.048 | 0.0535 | No | ||
| 47 | RPL39 | 12496 | 3657 | 0.042 | 0.0471 | No | ||
| 48 | PMM2 | 1670 1666 1707 22863 1756 | 3708 | 0.039 | 0.0448 | No | ||
| 49 | GHRL | 1183 17040 | 3710 | 0.039 | 0.0451 | No | ||
| 50 | EIF4H | 16352 | 3718 | 0.039 | 0.0451 | No | ||
| 51 | TNIP1 | 12192 20450 1320 | 3860 | 0.034 | 0.0377 | No | ||
| 52 | ALAS1 | 19013 | 3900 | 0.032 | 0.0359 | No | ||
| 53 | RPS11 | 11317 | 3922 | 0.031 | 0.0350 | No | ||
| 54 | FECH | 1975 23419 1964 | 4055 | 0.027 | 0.0281 | No | ||
| 55 | CYP11A1 | 4580 | 4164 | 0.024 | 0.0224 | No | ||
| 56 | EIF2C1 | 10672 | 4422 | 0.018 | 0.0086 | No | ||
| 57 | RPL13 | 11284 | 4430 | 0.018 | 0.0084 | No | ||
| 58 | PPOX | 13761 4104 | 4603 | 0.016 | -0.0009 | No | ||
| 59 | CYP39A1 | 1538 23232 | 4696 | 0.015 | -0.0057 | No | ||
| 60 | DHRS9 | 6308 2887 | 4721 | 0.014 | -0.0069 | No | ||
| 61 | CD28 | 14239 4092 | 4769 | 0.014 | -0.0093 | No | ||
| 62 | RPL5 | 9746 | 4800 | 0.014 | -0.0108 | No | ||
| 63 | MAT2B | 20494 1431 | 5321 | 0.010 | -0.0391 | No | ||
| 64 | RPS10 | 12464 | 5333 | 0.010 | -0.0396 | No | ||
| 65 | HSP90AA1 | 4883 4882 9131 | 5345 | 0.009 | -0.0401 | No | ||
| 66 | CYP7B1 | 15373 | 5353 | 0.009 | -0.0404 | No | ||
| 67 | TLR3 | 18884 | 5492 | 0.009 | -0.0478 | No | ||
| 68 | DDX25 | 19184 | 5505 | 0.009 | -0.0484 | No | ||
| 69 | ADORA2A | 8557 | 5522 | 0.008 | -0.0492 | No | ||
| 70 | PDZD3 | 19153 | 5526 | 0.008 | -0.0493 | No | ||
| 71 | TSC1 | 7235 | 5583 | 0.008 | -0.0523 | No | ||
| 72 | DCT | 4603 21723 | 5738 | 0.007 | -0.0606 | No | ||
| 73 | CYP11B2 | 24574 | 5812 | 0.007 | -0.0645 | No | ||
| 74 | INHA | 14214 | 5844 | 0.007 | -0.0661 | No | ||
| 75 | PAH | 19911 | 5984 | 0.006 | -0.0736 | No | ||
| 76 | RPL35 | 12360 | 6071 | 0.006 | -0.0783 | No | ||
| 77 | NQO1 | 18754 3836 | 6135 | 0.006 | -0.0816 | No | ||
| 78 | GLA | 4362 | 6220 | 0.005 | -0.0862 | No | ||
| 79 | CPOX | 1740 8780 | 6428 | 0.005 | -0.0974 | No | ||
| 80 | SAMD4A | 13211 | 6450 | 0.005 | -0.0985 | No | ||
| 81 | INHBB | 13858 | 6549 | 0.004 | -0.1038 | No | ||
| 82 | CHST8 | 12754 | 6587 | 0.004 | -0.1058 | No | ||
| 83 | RPS27 | 12183 | 6613 | 0.004 | -0.1071 | No | ||
| 84 | UGDH | 10249 | 6892 | 0.003 | -0.1222 | No | ||
| 85 | ABCA1 | 15881 | 6961 | 0.003 | -0.1259 | No | ||
| 86 | COQ3 | 16262 | 7183 | 0.003 | -0.1379 | No | ||
| 87 | CYP27B1 | 8831 3426 | 7202 | 0.003 | -0.1389 | No | ||
| 88 | BBOX1 | 5010 9290 | 7280 | 0.002 | -0.1430 | No | ||
| 89 | ADK | 3302 3454 8555 | 7303 | 0.002 | -0.1442 | No | ||
| 90 | BMP6 | 8659 | 7446 | 0.002 | -0.1519 | No | ||
| 91 | CHST9 | 7727 | 7669 | 0.001 | -0.1640 | No | ||
| 92 | TNFRSF8 | 2373 5784 | 7712 | 0.001 | -0.1663 | No | ||
| 93 | DEFB103A | 18665 | 8073 | 0.000 | -0.1859 | No | ||
| 94 | GCK | 20535 | 8153 | 0.000 | -0.1902 | No | ||
| 95 | MOCS2 | 21558 | 8214 | -0.000 | -0.1935 | No | ||
| 96 | PFKFB1 | 9552 | 8296 | -0.000 | -0.1979 | No | ||
| 97 | BACE2 | 1738 22687 1695 | 8391 | -0.001 | -0.2030 | No | ||
| 98 | GATM | 14451 | 8459 | -0.001 | -0.2066 | No | ||
| 99 | IL12B | 20918 | 8591 | -0.001 | -0.2138 | No | ||
| 100 | IL27 | 17636 | 8592 | -0.001 | -0.2137 | No | ||
| 101 | RPS3 | 6549 11295 | 8616 | -0.001 | -0.2150 | No | ||
| 102 | DAZL | 22944 | 8672 | -0.001 | -0.2180 | No | ||
| 103 | AGPAT1 | 23275 | 8905 | -0.002 | -0.2306 | No | ||
| 104 | UCN2 | 10308 | 8964 | -0.002 | -0.2337 | No | ||
| 105 | IL9 | 21444 | 9030 | -0.002 | -0.2373 | No | ||
| 106 | EIF4A2 | 4660 1679 1645 | 9078 | -0.002 | -0.2398 | No | ||
| 107 | MTRF1 | 3413 21949 | 9097 | -0.002 | -0.2408 | No | ||
| 108 | NDST1 | 23430 4885 | 9125 | -0.002 | -0.2422 | No | ||
| 109 | AK5 | 6042 | 9182 | -0.002 | -0.2452 | No | ||
| 110 | ALDH9A1 | 14064 | 9491 | -0.003 | -0.2620 | No | ||
| 111 | GLMN | 16452 3562 | 9611 | -0.003 | -0.2684 | No | ||
| 112 | CDO1 | 8731 | 9617 | -0.003 | -0.2687 | No | ||
| 113 | TLR9 | 19331 3127 | 9865 | -0.004 | -0.2821 | No | ||
| 114 | IL17F | 13992 | 9959 | -0.004 | -0.2871 | No | ||
| 115 | GPR143 | 24224 | 10591 | -0.006 | -0.3214 | No | ||
| 116 | HSD11B2 | 4872 | 10640 | -0.006 | -0.3240 | No | ||
| 117 | GRM8 | 4805 | 10783 | -0.006 | -0.3317 | No | ||
| 118 | PTGS1 | 2726 5316 15028 | 10857 | -0.007 | -0.3356 | No | ||
| 119 | TYR | 17763 | 10860 | -0.007 | -0.3356 | No | ||
| 120 | EREG | 4679 16797 | 11098 | -0.007 | -0.3485 | No | ||
| 121 | HS3ST3B1 | 7054 | 11143 | -0.008 | -0.3508 | No | ||
| 122 | EGFR | 1329 20944 | 11167 | -0.008 | -0.3520 | No | ||
| 123 | TLR8 | 9308 | 11176 | -0.008 | -0.3524 | No | ||
| 124 | RPS5 | 18391 | 11625 | -0.009 | -0.3767 | No | ||
| 125 | YBX2 | 20818 | 11868 | -0.011 | -0.3898 | No | ||
| 126 | PLCE1 | 13156 | 11989 | -0.011 | -0.3962 | No | ||
| 127 | EIF2AK4 | 14909 | 12182 | -0.012 | -0.4065 | No | ||
| 128 | YARS | 16071 | 12527 | -0.014 | -0.4251 | No | ||
| 129 | AKR1D1 | 17483 | 12732 | -0.015 | -0.4361 | No | ||
| 130 | PNPLA3 | 22406 | 12878 | -0.017 | -0.4439 | No | ||
| 131 | RPL26 | 9739 | 12927 | -0.017 | -0.4463 | No | ||
| 132 | PTGDS | 14658 | 12969 | -0.017 | -0.4484 | No | ||
| 133 | IL6 | 16895 | 13211 | -0.019 | -0.4613 | No | ||
| 134 | RPS6KB2 | 7198 | 13235 | -0.020 | -0.4624 | No | ||
| 135 | ALDH18A1 | 23682 | 13260 | -0.020 | -0.4635 | No | ||
| 136 | SFTPD | 21867 | 13329 | -0.020 | -0.4670 | No | ||
| 137 | AKT1 | 8568 | 13355 | -0.021 | -0.4682 | No | ||
| 138 | RPL18 | 450 5390 | 13376 | -0.021 | -0.4691 | No | ||
| 139 | IL4 | 9174 | 13402 | -0.021 | -0.4703 | No | ||
| 140 | ELA2 | 11925 8893 | 13684 | -0.025 | -0.4853 | No | ||
| 141 | LTA4H | 19906 3374 | 13686 | -0.025 | -0.4852 | No | ||
| 142 | BOLL | 7960 | 13722 | -0.026 | -0.4868 | No | ||
| 143 | TLR1 | 10190 | 13990 | -0.031 | -0.5011 | No | ||
| 144 | PEX7 | 19808 3305 | 14101 | -0.032 | -0.5068 | No | ||
| 145 | GCLC | 19374 | 14222 | -0.035 | -0.5130 | No | ||
| 146 | GHSR | 1810 15625 | 14369 | -0.038 | -0.5206 | No | ||
| 147 | CHST11 | 19921 | 14523 | -0.043 | -0.5285 | No | ||
| 148 | RPL41 | 12611 | 14629 | -0.046 | -0.5338 | No | ||
| 149 | TLR4 | 2329 10191 5770 | 14788 | -0.051 | -0.5420 | No | ||
| 150 | HPGD | 4867 | 14798 | -0.051 | -0.5420 | No | ||
| 151 | RPL37 | 12502 7421 22521 | 14998 | -0.061 | -0.5523 | No | ||
| 152 | SLC5A7 | 22938 | 15009 | -0.062 | -0.5523 | No | ||
| 153 | FOXP3 | 9804 5426 | 15013 | -0.062 | -0.5519 | No | ||
| 154 | HSD3B1 | 15227 | 15042 | -0.064 | -0.5528 | No | ||
| 155 | TGFB2 | 5743 | 15106 | -0.067 | -0.5556 | No | ||
| 156 | MAST2 | 9423 | 15200 | -0.073 | -0.5600 | No | ||
| 157 | PAIP1 | 21556 | 15276 | -0.078 | -0.5634 | No | ||
| 158 | RPL24 | 12664 | 15317 | -0.080 | -0.5648 | No | ||
| 159 | BRCA1 | 20213 | 15553 | -0.100 | -0.5767 | No | ||
| 160 | HSPB1 | 4879 | 15573 | -0.101 | -0.5768 | No | ||
| 161 | RPS17 | 9753 | 15582 | -0.102 | -0.5763 | No | ||
| 162 | RPL3 | 11330 | 15584 | -0.102 | -0.5754 | No | ||
| 163 | DGAT2 | 17748 | 15640 | -0.110 | -0.5774 | No | ||
| 164 | FARS2 | 21666 | 15647 | -0.110 | -0.5767 | No | ||
| 165 | PTS | 19126 | 15651 | -0.111 | -0.5758 | No | ||
| 166 | SPR | 17091 | 15712 | -0.117 | -0.5780 | No | ||
| 167 | TSTA3 | 22253 | 15775 | -0.125 | -0.5803 | No | ||
| 168 | APOA2 | 8615 4044 | 15777 | -0.125 | -0.5792 | No | ||
| 169 | EIF4G3 | 10517 | 15792 | -0.127 | -0.5788 | No | ||
| 170 | MTIF2 | 13354 | 15813 | -0.130 | -0.5786 | No | ||
| 171 | RPL8 | 22437 | 15856 | -0.136 | -0.5797 | No | ||
| 172 | NR5A1 | 14597 | 15857 | -0.136 | -0.5784 | No | ||
| 173 | RPL18A | 13358 | 15931 | -0.146 | -0.5810 | No | ||
| 174 | PTGES3 | 7101 12102 | 16038 | -0.165 | -0.5853 | No | ||
| 175 | PYCR1 | 20111 | 16064 | -0.168 | -0.5851 | No | ||
| 176 | PDSS1 | 15106 | 16150 | -0.186 | -0.5880 | No | ||
| 177 | RPL23A | 11193 | 16315 | -0.221 | -0.5949 | No | ||
| 178 | CHST12 | 16642 | 16398 | -0.242 | -0.5972 | No | ||
| 179 | BCAT1 | 1042 4434 | 16402 | -0.243 | -0.5951 | No | ||
| 180 | BCL3 | 8654 | 16492 | -0.266 | -0.5975 | No | ||
| 181 | ALAD | 15866 | 16542 | -0.279 | -0.5976 | Yes | ||
| 182 | SARS2 | 5 | 16573 | -0.287 | -0.5966 | Yes | ||
| 183 | ATF4 | 22417 2239 | 16600 | -0.293 | -0.5953 | Yes | ||
| 184 | RPL7A | 11312 | 16603 | -0.294 | -0.5927 | Yes | ||
| 185 | EEF1A1 | 38 38 8879 | 16614 | -0.296 | -0.5905 | Yes | ||
| 186 | PAICS | 16820 | 16646 | -0.307 | -0.5894 | Yes | ||
| 187 | RPS3A | 9755 | 16692 | -0.318 | -0.5889 | Yes | ||
| 188 | MRPL51 | 12361 7301 | 16699 | -0.319 | -0.5863 | Yes | ||
| 189 | ADCY7 | 8552 448 | 16725 | -0.325 | -0.5846 | Yes | ||
| 190 | EIF4G2 | 1908 8892 | 16813 | -0.350 | -0.5862 | Yes | ||
| 191 | SLC25A10 | 6572 | 16833 | -0.357 | -0.5839 | Yes | ||
| 192 | DEGS1 | 8850 | 16838 | -0.358 | -0.5808 | Yes | ||
| 193 | COX15 | 5931 | 16880 | -0.370 | -0.5797 | Yes | ||
| 194 | COPS5 | 4117 14002 | 16947 | -0.390 | -0.5797 | Yes | ||
| 195 | EIF4A3 | 9647 | 16988 | -0.404 | -0.5781 | Yes | ||
| 196 | EIF4B | 13279 7979 | 17024 | -0.419 | -0.5762 | Yes | ||
| 197 | RPL6 | 9747 | 17067 | -0.434 | -0.5744 | Yes | ||
| 198 | RPS19 | 5398 | 17101 | -0.445 | -0.5721 | Yes | ||
| 199 | ADM | 18125 | 17124 | -0.454 | -0.5692 | Yes | ||
| 200 | RPS2 | 9279 | 17144 | -0.459 | -0.5660 | Yes | ||
| 201 | NACA | 9444 5147 | 17147 | -0.459 | -0.5618 | Yes | ||
| 202 | HPRT1 | 2655 24339 408 | 17206 | -0.481 | -0.5606 | Yes | ||
| 203 | MRPL23 | 9738 | 17217 | -0.486 | -0.5566 | Yes | ||
| 204 | MRPL10 | 20681 | 17245 | -0.503 | -0.5535 | Yes | ||
| 205 | METAP1 | 15154 | 17267 | -0.511 | -0.5499 | Yes | ||
| 206 | COASY | 12953 | 17274 | -0.514 | -0.5455 | Yes | ||
| 207 | ACN9 | 12920 | 17291 | -0.525 | -0.5415 | Yes | ||
| 208 | RPL30 | 9742 | 17366 | -0.561 | -0.5404 | Yes | ||
| 209 | SCYE1 | 8897 | 17394 | -0.575 | -0.5366 | Yes | ||
| 210 | PET112L | 1824 15563 1819 | 17445 | -0.594 | -0.5338 | Yes | ||
| 211 | MRPL55 | 20864 | 17456 | -0.602 | -0.5288 | Yes | ||
| 212 | RPL31 | 14267 | 17485 | -0.621 | -0.5246 | Yes | ||
| 213 | IL10 | 14145 1510 1553 22902 | 17539 | -0.650 | -0.5215 | Yes | ||
| 214 | GMPS | 15578 | 17557 | -0.659 | -0.5164 | Yes | ||
| 215 | MRPS7 | 20595 | 17599 | -0.689 | -0.5122 | Yes | ||
| 216 | MRPL52 | 22019 | 17601 | -0.691 | -0.5059 | Yes | ||
| 217 | IRF4 | 21679 | 17618 | -0.695 | -0.5004 | Yes | ||
| 218 | UPF1 | 9718 18855 | 17675 | -0.729 | -0.4967 | Yes | ||
| 219 | EIF2AK1 | 4869 | 17694 | -0.738 | -0.4909 | Yes | ||
| 220 | DHPS | 3815 | 17728 | -0.760 | -0.4857 | Yes | ||
| 221 | MRPS24 | 12262 | 17776 | -0.799 | -0.4809 | Yes | ||
| 222 | KARS | 18738 | 17777 | -0.800 | -0.4735 | Yes | ||
| 223 | ABCF1 | 22996 10329 5897 | 17782 | -0.803 | -0.4664 | Yes | ||
| 224 | TYMS | 5810 5809 3606 3598 | 17793 | -0.813 | -0.4594 | Yes | ||
| 225 | RPL14 | 19267 | 17812 | -0.822 | -0.4528 | Yes | ||
| 226 | DPH1 | 4375 8580 8581 | 17821 | -0.831 | -0.4456 | Yes | ||
| 227 | EIF2B4 | 16574 | 17823 | -0.831 | -0.4380 | Yes | ||
| 228 | EIF2B5 | 1719 22822 | 17856 | -0.859 | -0.4318 | Yes | ||
| 229 | TUFM | 10608 | 17869 | -0.868 | -0.4245 | Yes | ||
| 230 | WARS | 2085 20984 | 17890 | -0.885 | -0.4174 | Yes | ||
| 231 | COX10 | 20408 | 17903 | -0.897 | -0.4098 | Yes | ||
| 232 | HSP90AB1 | 4880 9130 4881 | 17921 | -0.908 | -0.4023 | Yes | ||
| 233 | RPL21 | 9737 16624 5391 | 17936 | -0.918 | -0.3946 | Yes | ||
| 234 | RPL29 | 9741 | 17947 | -0.928 | -0.3866 | Yes | ||
| 235 | ETF1 | 23467 | 17951 | -0.929 | -0.3782 | Yes | ||
| 236 | GCLM | 9019 | 17961 | -0.934 | -0.3701 | Yes | ||
| 237 | PRG3 | 14971 | 18005 | -0.984 | -0.3634 | Yes | ||
| 238 | RPL4 | 7499 19411 | 18126 | -1.136 | -0.3595 | Yes | ||
| 239 | FARSB | 13909 | 18127 | -1.136 | -0.3490 | Yes | ||
| 240 | EIF2B1 | 16368 3458 | 18163 | -1.187 | -0.3400 | Yes | ||
| 241 | EIF1 | 23563 | 18209 | -1.270 | -0.3307 | Yes | ||
| 242 | RARS | 20496 | 18210 | -1.272 | -0.3190 | Yes | ||
| 243 | GUK1 | 20432 | 18229 | -1.307 | -0.3079 | Yes | ||
| 244 | MIF | 9387 | 18234 | -1.314 | -0.2961 | Yes | ||
| 245 | HRSP12 | 9123 9122 4870 9638 | 18279 | -1.407 | -0.2855 | Yes | ||
| 246 | UMPS | 22606 1760 | 18295 | -1.443 | -0.2730 | Yes | ||
| 247 | EIF5A | 11345 20379 6590 | 18300 | -1.448 | -0.2599 | Yes | ||
| 248 | PPAT | 6081 | 18307 | -1.467 | -0.2467 | Yes | ||
| 249 | DARS | 10375 13846 | 18330 | -1.506 | -0.2340 | Yes | ||
| 250 | EIF2B3 | 16118 | 18425 | -1.775 | -0.2228 | Yes | ||
| 251 | COQ7 | 17664 | 18455 | -1.875 | -0.2071 | Yes | ||
| 252 | RPL27A | 11181 6467 18130 | 18462 | -1.936 | -0.1896 | Yes | ||
| 253 | EIF5 | 5736 | 18479 | -2.027 | -0.1718 | Yes | ||
| 254 | MRPL3 | 3116 19335 | 18528 | -2.380 | -0.1525 | Yes | ||
| 255 | CHST7 | 24372 | 18553 | -2.751 | -0.1284 | Yes | ||
| 256 | CTPS | 2514 15772 | 18563 | -2.849 | -0.1026 | Yes | ||
| 257 | PABPC4 | 10510 2418 2548 | 18600 | -4.171 | -0.0662 | Yes | ||
| 258 | RPL38 | 12562 20606 7475 | 18616 | -7.271 | -0.0000 | Yes |