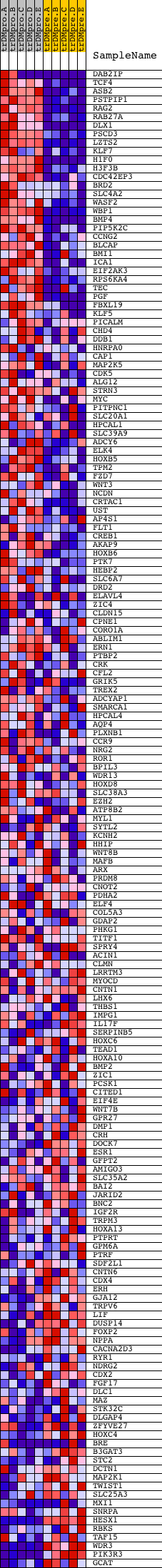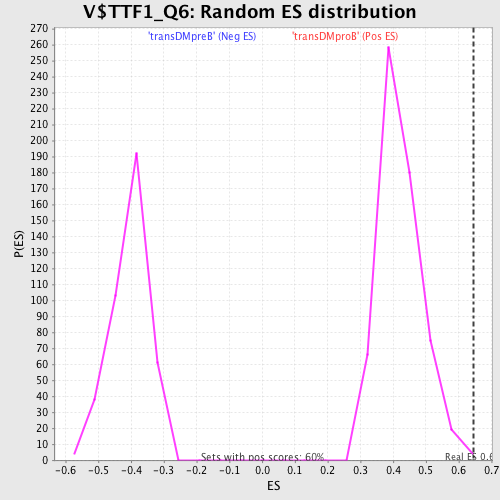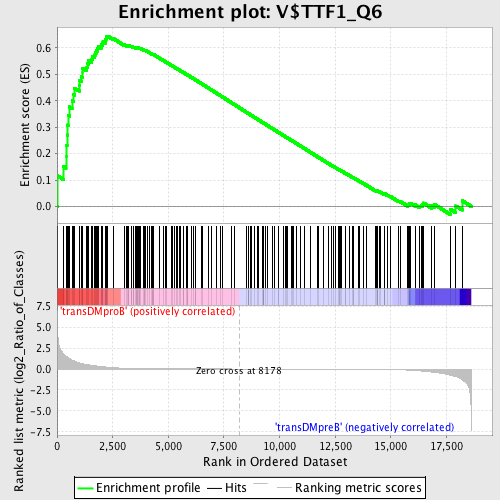
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$TTF1_Q6 |
| Enrichment Score (ES) | 0.6448716 |
| Normalized Enrichment Score (NES) | 1.5332742 |
| Nominal p-value | 0.0016611295 |
| FDR q-value | 0.07756682 |
| FWER p-Value | 0.631 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DAB2IP | 12808 2692 | 26 | 4.153 | 0.1157 | Yes | ||
| 2 | TCF4 | 5642 | 292 | 1.758 | 0.1509 | Yes | ||
| 3 | ASB2 | 20996 | 400 | 1.545 | 0.1886 | Yes | ||
| 4 | PSTPIP1 | 19437 | 404 | 1.541 | 0.2319 | Yes | ||
| 5 | RAG2 | 14941 | 450 | 1.456 | 0.2705 | Yes | ||
| 6 | RAB27A | 19381 | 477 | 1.391 | 0.3083 | Yes | ||
| 7 | DLX1 | 14988 | 502 | 1.333 | 0.3446 | Yes | ||
| 8 | PSCD3 | 16633 | 552 | 1.237 | 0.3768 | Yes | ||
| 9 | LZTS2 | 10362 | 674 | 1.057 | 0.4001 | Yes | ||
| 10 | KLF7 | 8205 | 734 | 0.981 | 0.4245 | Yes | ||
| 11 | H1F0 | 22428 | 790 | 0.929 | 0.4477 | Yes | ||
| 12 | H3F3B | 4839 | 993 | 0.739 | 0.4576 | Yes | ||
| 13 | CDC42EP3 | 6436 | 1007 | 0.722 | 0.4772 | Yes | ||
| 14 | BRD2 | 4735 8984 7628 | 1102 | 0.653 | 0.4906 | Yes | ||
| 15 | SLC4A2 | 9828 | 1151 | 0.617 | 0.5054 | Yes | ||
| 16 | WASF2 | 6326 | 1153 | 0.617 | 0.5227 | Yes | ||
| 17 | WBP1 | 17101 | 1336 | 0.531 | 0.5278 | Yes | ||
| 18 | BMP4 | 21863 | 1381 | 0.510 | 0.5398 | Yes | ||
| 19 | PIP5K2C | 19606 | 1398 | 0.504 | 0.5532 | Yes | ||
| 20 | CCNG2 | 16790 | 1554 | 0.436 | 0.5571 | Yes | ||
| 21 | BLCAP | 14371 | 1583 | 0.421 | 0.5674 | Yes | ||
| 22 | BMI1 | 4448 15113 | 1674 | 0.388 | 0.5735 | Yes | ||
| 23 | ICA1 | 17217 | 1709 | 0.375 | 0.5822 | Yes | ||
| 24 | EIF2AK3 | 17421 | 1750 | 0.362 | 0.5902 | Yes | ||
| 25 | RPS6KA4 | 23804 | 1799 | 0.345 | 0.5974 | Yes | ||
| 26 | TEC | 16514 | 1837 | 0.330 | 0.6046 | Yes | ||
| 27 | PGF | 21020 | 1976 | 0.293 | 0.6054 | Yes | ||
| 28 | FBXL19 | 6134 | 2005 | 0.283 | 0.6119 | Yes | ||
| 29 | KLF5 | 4456 | 2037 | 0.274 | 0.6179 | Yes | ||
| 30 | PICALM | 18191 | 2061 | 0.266 | 0.6242 | Yes | ||
| 31 | CHD4 | 8418 17281 4225 | 2169 | 0.239 | 0.6251 | Yes | ||
| 32 | DDB1 | 23931 | 2188 | 0.233 | 0.6307 | Yes | ||
| 33 | HNRPA0 | 13382 | 2199 | 0.230 | 0.6367 | Yes | ||
| 34 | CAP1 | 8684 | 2218 | 0.225 | 0.6420 | Yes | ||
| 35 | MAP2K5 | 19088 | 2276 | 0.211 | 0.6449 | Yes | ||
| 36 | CDK5 | 16591 | 2518 | 0.158 | 0.6363 | No | ||
| 37 | ALG12 | 22167 | 3012 | 0.089 | 0.6120 | No | ||
| 38 | STRN3 | 884 21074 | 3125 | 0.079 | 0.6082 | No | ||
| 39 | MYC | 22465 9435 | 3151 | 0.076 | 0.6090 | No | ||
| 40 | PITPNC1 | 7759 | 3222 | 0.069 | 0.6071 | No | ||
| 41 | SLC20A1 | 14855 | 3227 | 0.069 | 0.6089 | No | ||
| 42 | HPCAL1 | 21307 2123 | 3324 | 0.062 | 0.6054 | No | ||
| 43 | SLC39A9 | 11597 | 3328 | 0.061 | 0.6070 | No | ||
| 44 | ADCY6 | 22139 2283 8551 | 3431 | 0.054 | 0.6030 | No | ||
| 45 | ELK4 | 8895 4664 3937 | 3449 | 0.053 | 0.6035 | No | ||
| 46 | HOXB5 | 20687 | 3526 | 0.048 | 0.6008 | No | ||
| 47 | TPM2 | 2367 2437 | 3568 | 0.046 | 0.5998 | No | ||
| 48 | FZD7 | 14242 | 3580 | 0.045 | 0.6005 | No | ||
| 49 | WNT3 | 20635 | 3590 | 0.045 | 0.6013 | No | ||
| 50 | NCDN | 15756 2487 6471 | 3623 | 0.043 | 0.6008 | No | ||
| 51 | CRTAC1 | 23673 | 3654 | 0.042 | 0.6003 | No | ||
| 52 | UST | 6865 | 3685 | 0.040 | 0.5998 | No | ||
| 53 | AP4S1 | 21276 | 3707 | 0.039 | 0.5998 | No | ||
| 54 | FLT1 | 3483 16287 | 3749 | 0.038 | 0.5987 | No | ||
| 55 | CREB1 | 3990 8782 4558 4093 | 3899 | 0.032 | 0.5915 | No | ||
| 56 | AKAP9 | 16604 16603 4150 8303 | 3913 | 0.032 | 0.5917 | No | ||
| 57 | HOXB6 | 20688 | 3942 | 0.031 | 0.5910 | No | ||
| 58 | PTK7 | 22959 | 3970 | 0.030 | 0.5904 | No | ||
| 59 | HEBP2 | 19812 | 4067 | 0.027 | 0.5860 | No | ||
| 60 | SLC6A7 | 10772 | 4153 | 0.024 | 0.5820 | No | ||
| 61 | DRD2 | 19461 | 4246 | 0.022 | 0.5777 | No | ||
| 62 | ELAVL4 | 15805 4889 9137 | 4280 | 0.021 | 0.5765 | No | ||
| 63 | ZIC4 | 5992 | 4303 | 0.021 | 0.5759 | No | ||
| 64 | CLDN15 | 16667 3572 | 4337 | 0.020 | 0.5746 | No | ||
| 65 | CPNE1 | 11186 | 4352 | 0.020 | 0.5744 | No | ||
| 66 | CORO1A | 1066 17631 1074 | 4597 | 0.016 | 0.5617 | No | ||
| 67 | ABLIM1 | 3717 3766 3679 | 4778 | 0.014 | 0.5523 | No | ||
| 68 | ERN1 | 8137 | 4854 | 0.013 | 0.5486 | No | ||
| 69 | PTBP2 | 7074 | 4936 | 0.012 | 0.5445 | No | ||
| 70 | CRK | 4559 1249 | 5134 | 0.011 | 0.5342 | No | ||
| 71 | CFL2 | 2155 21069 | 5194 | 0.010 | 0.5313 | No | ||
| 72 | GRIK5 | 350 17930 | 5263 | 0.010 | 0.5278 | No | ||
| 73 | TREX2 | 24137 | 5368 | 0.009 | 0.5225 | No | ||
| 74 | ADCYAP1 | 4348 | 5388 | 0.009 | 0.5217 | No | ||
| 75 | SMARCA1 | 24165 | 5509 | 0.009 | 0.5154 | No | ||
| 76 | HPCAL4 | 16096 | 5562 | 0.008 | 0.5128 | No | ||
| 77 | AQP4 | 4403 | 5667 | 0.008 | 0.5074 | No | ||
| 78 | PLXNB1 | 19301 | 5673 | 0.008 | 0.5074 | No | ||
| 79 | CCR9 | 8753 | 5833 | 0.007 | 0.4989 | No | ||
| 80 | NRG2 | 4274 | 5851 | 0.007 | 0.4982 | No | ||
| 81 | ROR1 | 6472 | 5871 | 0.007 | 0.4974 | No | ||
| 82 | BPIL3 | 10459 | 6035 | 0.006 | 0.4887 | No | ||
| 83 | WDR13 | 7859 13112 | 6146 | 0.006 | 0.4829 | No | ||
| 84 | HOXD8 | 9116 | 6228 | 0.005 | 0.4787 | No | ||
| 85 | SLC38A3 | 13319 | 6240 | 0.005 | 0.4782 | No | ||
| 86 | EZH2 | 1092 17163 | 6481 | 0.005 | 0.4653 | No | ||
| 87 | ATP8B2 | 12055 | 6543 | 0.004 | 0.4622 | No | ||
| 88 | MYL1 | 4015 | 6797 | 0.004 | 0.4485 | No | ||
| 89 | SYTL2 | 18190 1539 3730 | 6956 | 0.003 | 0.4401 | No | ||
| 90 | KCNH2 | 16592 | 7169 | 0.003 | 0.4286 | No | ||
| 91 | HHIP | 4849 | 7361 | 0.002 | 0.4183 | No | ||
| 92 | WNT8B | 23841 | 7443 | 0.002 | 0.4140 | No | ||
| 93 | MAFB | 14367 | 7840 | 0.001 | 0.3926 | No | ||
| 94 | ARX | 24290 | 7973 | 0.001 | 0.3854 | No | ||
| 95 | PRDM8 | 8084 | 8501 | -0.001 | 0.3569 | No | ||
| 96 | CNOT2 | 12992 3332 453 3357 | 8583 | -0.001 | 0.3525 | No | ||
| 97 | PDHA2 | 15153 | 8588 | -0.001 | 0.3523 | No | ||
| 98 | ELF4 | 24162 | 8590 | -0.001 | 0.3523 | No | ||
| 99 | COL5A3 | 19219 | 8698 | -0.001 | 0.3465 | No | ||
| 100 | GDAP2 | 15481 | 8735 | -0.001 | 0.3446 | No | ||
| 101 | PHKG1 | 16361 | 8881 | -0.002 | 0.3368 | No | ||
| 102 | TITF1 | 10180 21062 | 9011 | -0.002 | 0.3298 | No | ||
| 103 | SPRY4 | 23448 | 9072 | -0.002 | 0.3266 | No | ||
| 104 | ACIN1 | 3319 2807 21832 | 9219 | -0.002 | 0.3188 | No | ||
| 105 | CLMN | 20988 2119 2104 | 9262 | -0.002 | 0.3166 | No | ||
| 106 | LRRTM3 | 19744 | 9277 | -0.003 | 0.3159 | No | ||
| 107 | MYOCD | 1382 20406 1401 | 9279 | -0.003 | 0.3159 | No | ||
| 108 | CNTN1 | 4538 | 9385 | -0.003 | 0.3103 | No | ||
| 109 | LHX6 | 9275 2914 | 9464 | -0.003 | 0.3062 | No | ||
| 110 | THBS1 | 5748 14910 | 9687 | -0.003 | 0.2942 | No | ||
| 111 | IMPG1 | 19052 | 9772 | -0.004 | 0.2898 | No | ||
| 112 | IL17F | 13992 | 9959 | -0.004 | 0.2798 | No | ||
| 113 | SERPINB5 | 9861 | 9961 | -0.004 | 0.2799 | No | ||
| 114 | HOXC6 | 22339 2302 | 10191 | -0.005 | 0.2676 | No | ||
| 115 | TEAD1 | 18121 | 10267 | -0.005 | 0.2636 | No | ||
| 116 | HOXA10 | 17145 989 | 10319 | -0.005 | 0.2610 | No | ||
| 117 | BMP2 | 14833 | 10344 | -0.005 | 0.2599 | No | ||
| 118 | ZIC1 | 19040 | 10535 | -0.006 | 0.2497 | No | ||
| 119 | PCSK1 | 21600 | 10560 | -0.006 | 0.2486 | No | ||
| 120 | CITED1 | 24091 | 10637 | -0.006 | 0.2446 | No | ||
| 121 | EIF4E | 15403 1827 8890 | 10775 | -0.006 | 0.2374 | No | ||
| 122 | WNT7B | 22172 | 10957 | -0.007 | 0.2278 | No | ||
| 123 | GPR27 | 13703 | 11112 | -0.008 | 0.2196 | No | ||
| 124 | DMP1 | 16776 | 11374 | -0.008 | 0.2057 | No | ||
| 125 | CRH | 8784 | 11723 | -0.010 | 0.1871 | No | ||
| 126 | DOCK7 | 12506 | 11728 | -0.010 | 0.1872 | No | ||
| 127 | ESR1 | 20097 4685 | 11982 | -0.011 | 0.1738 | No | ||
| 128 | GFPT2 | 4775 | 12205 | -0.012 | 0.1621 | No | ||
| 129 | AMIGO3 | 19314 | 12323 | -0.013 | 0.1561 | No | ||
| 130 | SLC35A2 | 24389 | 12437 | -0.014 | 0.1504 | No | ||
| 131 | BAI2 | 2541 2379 16067 | 12521 | -0.014 | 0.1463 | No | ||
| 132 | JARID2 | 9200 | 12634 | -0.015 | 0.1406 | No | ||
| 133 | BNC2 | 15850 | 12671 | -0.015 | 0.1391 | No | ||
| 134 | IGF2R | 23123 | 12696 | -0.015 | 0.1382 | No | ||
| 135 | TRPM3 | 23904 10357 5927 | 12743 | -0.016 | 0.1361 | No | ||
| 136 | HOXA13 | 9107 | 12778 | -0.016 | 0.1347 | No | ||
| 137 | PTPRT | 5333 | 12947 | -0.017 | 0.1261 | No | ||
| 138 | GPM6A | 3834 10618 | 12956 | -0.017 | 0.1262 | No | ||
| 139 | PTRF | 9667 | 13144 | -0.019 | 0.1166 | No | ||
| 140 | SDF2L1 | 22649 | 13294 | -0.020 | 0.1091 | No | ||
| 141 | CNTN6 | 17346 | 13319 | -0.020 | 0.1083 | No | ||
| 142 | CDX4 | 24273 | 13337 | -0.021 | 0.1080 | No | ||
| 143 | ERH | 9614 4681 | 13528 | -0.023 | 0.0983 | No | ||
| 144 | GJA12 | 20433 1323 | 13609 | -0.024 | 0.0947 | No | ||
| 145 | TRPV6 | 17171 | 13767 | -0.026 | 0.0869 | No | ||
| 146 | LIF | 956 667 20960 | 13887 | -0.028 | 0.0812 | No | ||
| 147 | DUSP14 | 12124 | 14289 | -0.036 | 0.0605 | No | ||
| 148 | FOXP2 | 4312 17523 8523 | 14370 | -0.039 | 0.0573 | No | ||
| 149 | NPPA | 298 | 14382 | -0.039 | 0.0578 | No | ||
| 150 | CACNA2D3 | 8677 | 14394 | -0.039 | 0.0583 | No | ||
| 151 | RYR1 | 17903 17902 9770 | 14405 | -0.039 | 0.0588 | No | ||
| 152 | NDRG2 | 21843 | 14483 | -0.041 | 0.0558 | No | ||
| 153 | CDX2 | 16289 | 14529 | -0.043 | 0.0546 | No | ||
| 154 | FGF17 | 21757 | 14703 | -0.048 | 0.0466 | No | ||
| 155 | DLC1 | 11931 3855 | 14705 | -0.048 | 0.0479 | No | ||
| 156 | MAZ | 1327 17623 | 14733 | -0.049 | 0.0478 | No | ||
| 157 | STK32C | 17580 | 14860 | -0.054 | 0.0425 | No | ||
| 158 | DLGAP4 | 2950 10461 | 14989 | -0.060 | 0.0373 | No | ||
| 159 | ZFYVE27 | 23849 | 15366 | -0.083 | 0.0192 | No | ||
| 160 | HOXC4 | 4864 | 15454 | -0.090 | 0.0170 | No | ||
| 161 | BRE | 3640 4226 16879 | 15768 | -0.124 | 0.0036 | No | ||
| 162 | B3GAT3 | 23938 | 15796 | -0.128 | 0.0057 | No | ||
| 163 | STC2 | 5526 | 15820 | -0.131 | 0.0081 | No | ||
| 164 | DCTN1 | 1100 17392 | 15839 | -0.134 | 0.0109 | No | ||
| 165 | MAP2K1 | 19082 | 15878 | -0.140 | 0.0128 | No | ||
| 166 | TWIST1 | 21291 | 16092 | -0.175 | 0.0062 | No | ||
| 167 | SLC25A3 | 19644 | 16308 | -0.219 | 0.0007 | No | ||
| 168 | MXI1 | 23813 5139 | 16361 | -0.231 | 0.0044 | No | ||
| 169 | SNRPA | 7007 11992 7008 | 16440 | -0.252 | 0.0073 | No | ||
| 170 | HESX1 | 22070 | 16470 | -0.259 | 0.0130 | No | ||
| 171 | RBKS | 16568 | 16828 | -0.354 | 0.0036 | No | ||
| 172 | TAF15 | 20731 | 16976 | -0.399 | 0.0069 | No | ||
| 173 | WDR3 | 15225 | 17668 | -0.722 | -0.0102 | No | ||
| 174 | PIK3R3 | 5248 | 17924 | -0.910 | 0.0017 | No | ||
| 175 | GCAT | 11234 | 18208 | -1.270 | 0.0221 | No |