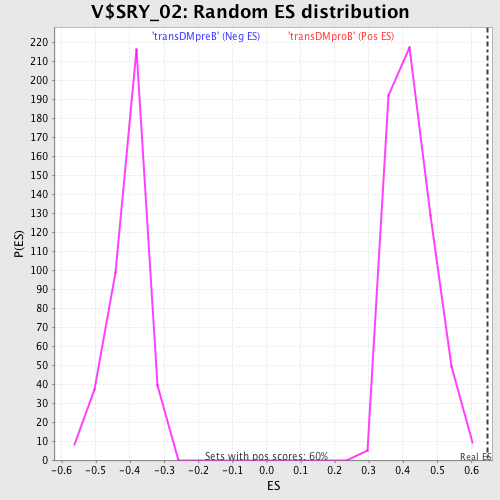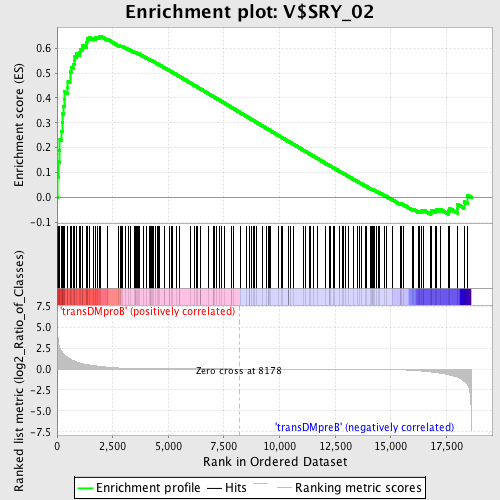
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$SRY_02 |
| Enrichment Score (ES) | 0.64891803 |
| Normalized Enrichment Score (NES) | 1.5322931 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0748294 |
| FWER p-Value | 0.64 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DUSP6 | 19891 3399 | 15 | 4.550 | 0.0829 | Yes | ||
| 2 | SATB1 | 5408 | 53 | 3.347 | 0.1425 | Yes | ||
| 3 | DDX6 | 3086 8844 | 98 | 2.683 | 0.1895 | Yes | ||
| 4 | TIAM1 | 22547 | 127 | 2.451 | 0.2331 | Yes | ||
| 5 | MAML1 | 20477 | 209 | 2.068 | 0.2668 | Yes | ||
| 6 | NFKBIE | 23225 1556 | 225 | 2.015 | 0.3030 | Yes | ||
| 7 | KLF3 | 4961 | 257 | 1.890 | 0.3361 | Yes | ||
| 8 | TCF4 | 5642 | 292 | 1.758 | 0.3666 | Yes | ||
| 9 | RAPGEF5 | 5739 10155 21123 | 333 | 1.657 | 0.3949 | Yes | ||
| 10 | PIK3CG | 6635 | 341 | 1.639 | 0.4247 | Yes | ||
| 11 | MBNL2 | 21932 | 484 | 1.376 | 0.4424 | Yes | ||
| 12 | MYLK | 22778 4213 | 488 | 1.366 | 0.4673 | Yes | ||
| 13 | HSD11B1 | 9125 4019 | 581 | 1.194 | 0.4843 | Yes | ||
| 14 | NFIX | 9458 | 582 | 1.194 | 0.5063 | Yes | ||
| 15 | H2AFY | 21447 | 637 | 1.098 | 0.5236 | Yes | ||
| 16 | KLF7 | 8205 | 734 | 0.981 | 0.5364 | Yes | ||
| 17 | SMAD3 | 19084 | 774 | 0.945 | 0.5517 | Yes | ||
| 18 | COMMD3 | 73 | 800 | 0.916 | 0.5672 | Yes | ||
| 19 | TNFRSF19 | 21799 3033 | 875 | 0.850 | 0.5788 | Yes | ||
| 20 | CDC42EP3 | 6436 | 1007 | 0.722 | 0.5850 | Yes | ||
| 21 | GATA2 | 17370 | 1067 | 0.669 | 0.5941 | Yes | ||
| 22 | ICK | 7135 12146 | 1150 | 0.618 | 0.6010 | Yes | ||
| 23 | EMCN | 7211 | 1156 | 0.615 | 0.6121 | Yes | ||
| 24 | PPM1D | 20721 | 1315 | 0.540 | 0.6135 | Yes | ||
| 25 | PIAS1 | 7126 | 1335 | 0.531 | 0.6222 | Yes | ||
| 26 | WASL | 17203 | 1346 | 0.526 | 0.6313 | Yes | ||
| 27 | LDB1 | 23654 | 1350 | 0.523 | 0.6408 | Yes | ||
| 28 | MSI2 | 8030 1268 | 1451 | 0.482 | 0.6443 | Yes | ||
| 29 | ZFYVE1 | 10138 | 1657 | 0.395 | 0.6404 | Yes | ||
| 30 | KBTBD2 | 17137 | 1731 | 0.368 | 0.6432 | Yes | ||
| 31 | MYO18A | 20766 | 1818 | 0.336 | 0.6448 | Yes | ||
| 32 | SFRS7 | 22889 | 1898 | 0.314 | 0.6463 | Yes | ||
| 33 | NFE2L1 | 9457 | 1952 | 0.301 | 0.6489 | Yes | ||
| 34 | ZADH2 | 23501 | 2250 | 0.217 | 0.6368 | No | ||
| 35 | TCF15 | 14798 | 2767 | 0.118 | 0.6110 | No | ||
| 36 | PAK1 | 9527 | 2868 | 0.104 | 0.6075 | No | ||
| 37 | ULK1 | 16436 | 2912 | 0.099 | 0.6070 | No | ||
| 38 | TBX6 | 1993 18075 | 2940 | 0.096 | 0.6073 | No | ||
| 39 | NFIA | 16172 5170 | 3093 | 0.082 | 0.6006 | No | ||
| 40 | FGFR2 | 1917 4722 1119 | 3186 | 0.073 | 0.5969 | No | ||
| 41 | RHOBTB1 | 12789 | 3302 | 0.063 | 0.5918 | No | ||
| 42 | CAMK1G | 13710 4129 | 3483 | 0.051 | 0.5830 | No | ||
| 43 | SLC26A9 | 11560 980 4067 | 3510 | 0.049 | 0.5825 | No | ||
| 44 | HOXB5 | 20687 | 3526 | 0.048 | 0.5826 | No | ||
| 45 | NSF | 20192 | 3565 | 0.046 | 0.5814 | No | ||
| 46 | DHRS3 | 15997 | 3601 | 0.044 | 0.5803 | No | ||
| 47 | SOX5 | 9850 1044 5480 16937 | 3677 | 0.041 | 0.5769 | No | ||
| 48 | CTLA4 | 14238 | 3682 | 0.040 | 0.5775 | No | ||
| 49 | OLFM1 | 2918 2971 | 3870 | 0.033 | 0.5680 | No | ||
| 50 | LRCH2 | 24035 | 3997 | 0.029 | 0.5617 | No | ||
| 51 | ANGPT1 | 8588 2260 22303 | 4133 | 0.025 | 0.5548 | No | ||
| 52 | PELI2 | 8217 | 4180 | 0.024 | 0.5528 | No | ||
| 53 | APBA1 | 11483 | 4205 | 0.023 | 0.5519 | No | ||
| 54 | HS3ST1 | 16542 | 4232 | 0.022 | 0.5509 | No | ||
| 55 | ELAVL4 | 15805 4889 9137 | 4280 | 0.021 | 0.5487 | No | ||
| 56 | CPNE1 | 11186 | 4352 | 0.020 | 0.5452 | No | ||
| 57 | FOXB1 | 12254 19069 | 4427 | 0.018 | 0.5416 | No | ||
| 58 | TCF1 | 16416 | 4523 | 0.017 | 0.5367 | No | ||
| 59 | GPR110 | 23234 | 4550 | 0.016 | 0.5356 | No | ||
| 60 | CACNA1E | 4465 | 4619 | 0.016 | 0.5322 | No | ||
| 61 | SLC13A1 | 17205 | 4808 | 0.013 | 0.5223 | No | ||
| 62 | IL5RA | 17051 | 4819 | 0.013 | 0.5220 | No | ||
| 63 | FN1 | 4091 4094 971 13929 | 5066 | 0.011 | 0.5088 | No | ||
| 64 | LPHN2 | 13619 | 5067 | 0.011 | 0.5091 | No | ||
| 65 | PIK3R1 | 3170 | 5127 | 0.011 | 0.5061 | No | ||
| 66 | CFL2 | 2155 21069 | 5194 | 0.010 | 0.5027 | No | ||
| 67 | MITF | 17349 | 5365 | 0.009 | 0.4936 | No | ||
| 68 | FGF9 | 8966 | 5506 | 0.009 | 0.4862 | No | ||
| 69 | PAK3 | 9528 | 5514 | 0.008 | 0.4860 | No | ||
| 70 | GNAO1 | 4785 3829 | 6003 | 0.006 | 0.4596 | No | ||
| 71 | MAB21L1 | 15589 5046 | 6183 | 0.006 | 0.4500 | No | ||
| 72 | CHN1 | 4246 | 6272 | 0.005 | 0.4454 | No | ||
| 73 | QKI | 23128 5340 | 6318 | 0.005 | 0.4430 | No | ||
| 74 | LRRN1 | 17343 | 6447 | 0.005 | 0.4362 | No | ||
| 75 | OLFML3 | 15216 | 6451 | 0.005 | 0.4361 | No | ||
| 76 | BAMBI | 23629 | 6458 | 0.005 | 0.4359 | No | ||
| 77 | PTN | 5323 | 6813 | 0.004 | 0.4167 | No | ||
| 78 | DLL1 | 23119 | 7020 | 0.003 | 0.4056 | No | ||
| 79 | NFIB | 15855 | 7045 | 0.003 | 0.4044 | No | ||
| 80 | ARHGAP15 | 8004 377 13310 | 7076 | 0.003 | 0.4028 | No | ||
| 81 | MRC2 | 1386 5117 | 7153 | 0.003 | 0.3987 | No | ||
| 82 | HOXD10 | 14983 | 7290 | 0.002 | 0.3914 | No | ||
| 83 | MLLT7 | 24278 | 7398 | 0.002 | 0.3856 | No | ||
| 84 | NKX2-2 | 5176 | 7544 | 0.002 | 0.3778 | No | ||
| 85 | PTGFRN | 15224 | 7823 | 0.001 | 0.3628 | No | ||
| 86 | ZIC2 | 21927 | 7847 | 0.001 | 0.3615 | No | ||
| 87 | LEMD1 | 10023 537 | 7917 | 0.001 | 0.3578 | No | ||
| 88 | EPHA7 | 8909 4674 | 8260 | -0.000 | 0.3393 | No | ||
| 89 | PCF11 | 121 | 8517 | -0.001 | 0.3254 | No | ||
| 90 | CCNJ | 23859 | 8644 | -0.001 | 0.3186 | No | ||
| 91 | NR4A1 | 9099 | 8658 | -0.001 | 0.3179 | No | ||
| 92 | PURA | 9670 | 8758 | -0.001 | 0.3126 | No | ||
| 93 | NUDT11 | 24387 | 8806 | -0.001 | 0.3100 | No | ||
| 94 | RBM9 | 2276 13528 | 8850 | -0.002 | 0.3077 | No | ||
| 95 | ESRRG | 6447 | 8980 | -0.002 | 0.3008 | No | ||
| 96 | KERA | 19895 | 9216 | -0.002 | 0.2881 | No | ||
| 97 | MID2 | 24232 | 9405 | -0.003 | 0.2779 | No | ||
| 98 | NR3C2 | 18562 13798 | 9488 | -0.003 | 0.2736 | No | ||
| 99 | FST | 8985 | 9508 | -0.003 | 0.2726 | No | ||
| 100 | TACSTD2 | 17126 | 9532 | -0.003 | 0.2714 | No | ||
| 101 | EBF2 | 21975 | 9602 | -0.003 | 0.2677 | No | ||
| 102 | RCN1 | 9710 | 9944 | -0.004 | 0.2493 | No | ||
| 103 | EN2 | 16898 | 10072 | -0.004 | 0.2425 | No | ||
| 104 | KCNN2 | 8933 1949 | 10148 | -0.005 | 0.2385 | No | ||
| 105 | HEY1 | 15378 | 10378 | -0.005 | 0.2262 | No | ||
| 106 | B3GALT2 | 6498 | 10391 | -0.005 | 0.2257 | No | ||
| 107 | DTNA | 8870 4647 23611 1991 | 10399 | -0.005 | 0.2254 | No | ||
| 108 | DEPDC2 | 14295 | 10417 | -0.005 | 0.2246 | No | ||
| 109 | MLLT3 | 2359 15847 2413 | 10499 | -0.006 | 0.2203 | No | ||
| 110 | ELAVL2 | 15839 | 10643 | -0.006 | 0.2126 | No | ||
| 111 | LRP5 | 23948 9285 | 11079 | -0.007 | 0.1892 | No | ||
| 112 | LRP1 | 9284 | 11148 | -0.008 | 0.1856 | No | ||
| 113 | TIAL1 | 996 5753 | 11345 | -0.008 | 0.1752 | No | ||
| 114 | SIX4 | 5445 | 11383 | -0.008 | 0.1733 | No | ||
| 115 | A2BP1 | 11212 1652 1689 | 11544 | -0.009 | 0.1648 | No | ||
| 116 | CRH | 8784 | 11723 | -0.010 | 0.1553 | No | ||
| 117 | SOX6 | 3684 5481 9851 | 12076 | -0.012 | 0.1365 | No | ||
| 118 | KLHL13 | 12536 | 12221 | -0.012 | 0.1289 | No | ||
| 119 | CLDN18 | 19028 | 12242 | -0.012 | 0.1280 | No | ||
| 120 | POU4F1 | 21727 | 12305 | -0.013 | 0.1249 | No | ||
| 121 | GNAQ | 4786 23909 3685 | 12410 | -0.013 | 0.1195 | No | ||
| 122 | NR4A3 | 9473 16212 5183 | 12466 | -0.014 | 0.1168 | No | ||
| 123 | BNC2 | 15850 | 12671 | -0.015 | 0.1060 | No | ||
| 124 | CD36 | 8712 | 12827 | -0.016 | 0.0979 | No | ||
| 125 | SEMA6C | 1797 5424 | 12858 | -0.016 | 0.0966 | No | ||
| 126 | ABCA6 | 1470 20165 | 12978 | -0.017 | 0.0905 | No | ||
| 127 | MOBP | 5112 | 13112 | -0.018 | 0.0836 | No | ||
| 128 | SLC7A1 | 4430 16285 | 13328 | -0.020 | 0.0723 | No | ||
| 129 | TGFB3 | 10161 | 13520 | -0.023 | 0.0624 | No | ||
| 130 | FBN2 | 23433 | 13570 | -0.024 | 0.0602 | No | ||
| 131 | NTF3 | 16991 | 13671 | -0.025 | 0.0552 | No | ||
| 132 | SOX2 | 9849 15612 | 13859 | -0.028 | 0.0456 | No | ||
| 133 | TECTA | 19159 | 13884 | -0.028 | 0.0448 | No | ||
| 134 | AP1G2 | 2677 21827 | 14069 | -0.032 | 0.0354 | No | ||
| 135 | STC1 | 21974 | 14151 | -0.033 | 0.0316 | No | ||
| 136 | ALDH1A2 | 19386 | 14170 | -0.034 | 0.0313 | No | ||
| 137 | TBX5 | 3531 5634 | 14175 | -0.034 | 0.0317 | No | ||
| 138 | XKRX | 301 | 14206 | -0.035 | 0.0307 | No | ||
| 139 | STAG2 | 5521 | 14273 | -0.036 | 0.0278 | No | ||
| 140 | FOXP2 | 4312 17523 8523 | 14370 | -0.039 | 0.0233 | No | ||
| 141 | SP8 | 21122 | 14452 | -0.041 | 0.0196 | No | ||
| 142 | TNFRSF19L | 11498 6727 6726 | 14481 | -0.041 | 0.0189 | No | ||
| 143 | PHF16 | 11795 6901 | 14698 | -0.048 | 0.0081 | No | ||
| 144 | POU3F4 | 347 | 14812 | -0.052 | 0.0029 | No | ||
| 145 | AP4M1 | 3499 16656 | 15074 | -0.065 | -0.0101 | No | ||
| 146 | RBBP4 | 5364 | 15445 | -0.089 | -0.0285 | No | ||
| 147 | HOXC4 | 4864 | 15454 | -0.090 | -0.0272 | No | ||
| 148 | SLC12A8 | 5059 | 15461 | -0.091 | -0.0259 | No | ||
| 149 | MDGA1 | 13231 | 15570 | -0.101 | -0.0299 | No | ||
| 150 | MYST3 | 18651 | 15979 | -0.156 | -0.0491 | No | ||
| 151 | DHX40 | 20307 | 16009 | -0.161 | -0.0477 | No | ||
| 152 | CSRP2BP | 2696 10458 | 16243 | -0.205 | -0.0566 | No | ||
| 153 | PUM1 | 8160 | 16302 | -0.218 | -0.0557 | No | ||
| 154 | MXI1 | 23813 5139 | 16361 | -0.231 | -0.0546 | No | ||
| 155 | HLX1 | 9093 | 16446 | -0.254 | -0.0545 | No | ||
| 156 | HESX1 | 22070 | 16470 | -0.259 | -0.0510 | No | ||
| 157 | CCBL1 | 14630 | 16801 | -0.346 | -0.0625 | No | ||
| 158 | EIF4G2 | 1908 8892 | 16813 | -0.350 | -0.0567 | No | ||
| 159 | YWHAG | 16339 | 16844 | -0.360 | -0.0517 | No | ||
| 160 | PRKAG1 | 22135 | 16997 | -0.407 | -0.0524 | No | ||
| 161 | NANOS1 | 6858 | 17066 | -0.434 | -0.0481 | No | ||
| 162 | GRAP | 20851 | 17238 | -0.499 | -0.0482 | No | ||
| 163 | MPP6 | 17448 | 17589 | -0.682 | -0.0546 | No | ||
| 164 | E2F1 | 14384 | 17648 | -0.711 | -0.0447 | No | ||
| 165 | CHD1 | 23377 | 17988 | -0.962 | -0.0454 | No | ||
| 166 | LMO4 | 15151 | 18004 | -0.982 | -0.0281 | No | ||
| 167 | RBM8A | 12239 7212 | 18290 | -1.435 | -0.0172 | No | ||
| 168 | MCM7 | 9372 3568 | 18458 | -1.891 | 0.0086 | No |