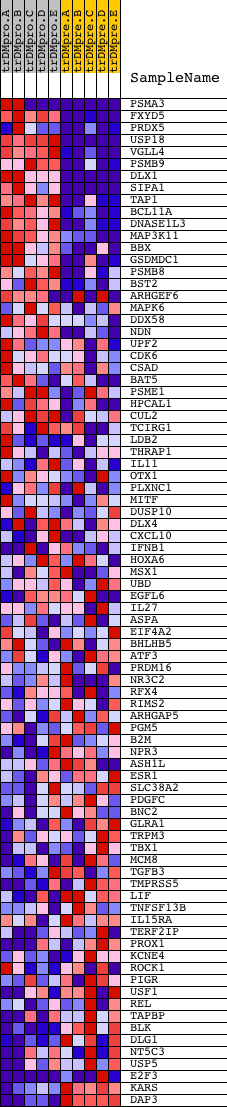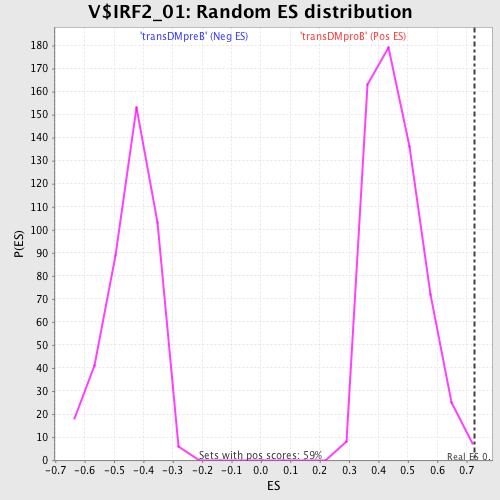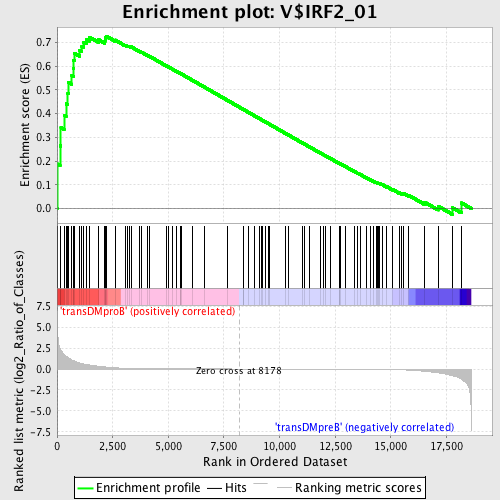
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$IRF2_01 |
| Enrichment Score (ES) | 0.7272368 |
| Normalized Enrichment Score (NES) | 1.5822656 |
| Nominal p-value | 0.006779661 |
| FDR q-value | 0.050833832 |
| FWER p-Value | 0.318 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA3 | 9632 5298 | 7 | 5.394 | 0.1880 | Yes | ||
| 2 | FXYD5 | 17876 2102 | 134 | 2.407 | 0.2653 | Yes | ||
| 3 | PRDX5 | 7053 3732 | 172 | 2.242 | 0.3417 | Yes | ||
| 4 | USP18 | 17301 | 316 | 1.700 | 0.3933 | Yes | ||
| 5 | VGLL4 | 10558 | 406 | 1.537 | 0.4422 | Yes | ||
| 6 | PSMB9 | 23021 | 482 | 1.379 | 0.4864 | Yes | ||
| 7 | DLX1 | 14988 | 502 | 1.333 | 0.5319 | Yes | ||
| 8 | SIPA1 | 9820 | 661 | 1.072 | 0.5608 | Yes | ||
| 9 | TAP1 | 23283 | 736 | 0.979 | 0.5910 | Yes | ||
| 10 | BCL11A | 4691 | 748 | 0.967 | 0.6242 | Yes | ||
| 11 | DNASE1L3 | 21925 | 779 | 0.938 | 0.6554 | Yes | ||
| 12 | MAP3K11 | 11163 | 1013 | 0.714 | 0.6678 | Yes | ||
| 13 | BBX | 7685 | 1111 | 0.646 | 0.6851 | Yes | ||
| 14 | GSDMDC1 | 22453 12780 | 1199 | 0.592 | 0.7011 | Yes | ||
| 15 | PSMB8 | 5000 | 1331 | 0.532 | 0.7126 | Yes | ||
| 16 | BST2 | 18845 | 1449 | 0.483 | 0.7232 | Yes | ||
| 17 | ARHGEF6 | 13297 13104 | 1859 | 0.324 | 0.7124 | Yes | ||
| 18 | MAPK6 | 19062 | 2145 | 0.244 | 0.7056 | Yes | ||
| 19 | DDX58 | 6043 | 2155 | 0.241 | 0.7135 | Yes | ||
| 20 | NDN | 18222 | 2163 | 0.240 | 0.7215 | Yes | ||
| 21 | UPF2 | 11591 | 2205 | 0.227 | 0.7272 | Yes | ||
| 22 | CDK6 | 16600 | 2602 | 0.144 | 0.7109 | No | ||
| 23 | CSAD | 10899 | 3059 | 0.085 | 0.6893 | No | ||
| 24 | BAT5 | 23261 | 3183 | 0.073 | 0.6852 | No | ||
| 25 | PSME1 | 5300 9637 | 3241 | 0.068 | 0.6845 | No | ||
| 26 | HPCAL1 | 21307 2123 | 3324 | 0.062 | 0.6822 | No | ||
| 27 | CUL2 | 23630 | 3714 | 0.039 | 0.6626 | No | ||
| 28 | TCIRG1 | 3700 3752 23951 | 3790 | 0.036 | 0.6598 | No | ||
| 29 | LDB2 | 16538 4989 | 4078 | 0.027 | 0.6453 | No | ||
| 30 | THRAP1 | 20309 | 4157 | 0.024 | 0.6419 | No | ||
| 31 | IL11 | 17981 | 4921 | 0.012 | 0.6012 | No | ||
| 32 | OTX1 | 20517 | 5019 | 0.012 | 0.5963 | No | ||
| 33 | PLXNC1 | 7056 | 5171 | 0.011 | 0.5886 | No | ||
| 34 | MITF | 17349 | 5365 | 0.009 | 0.5785 | No | ||
| 35 | DUSP10 | 4003 14016 | 5533 | 0.008 | 0.5698 | No | ||
| 36 | DLX4 | 20282 1225 | 5566 | 0.008 | 0.5683 | No | ||
| 37 | CXCL10 | 16477 | 5591 | 0.008 | 0.5673 | No | ||
| 38 | IFNB1 | 15846 | 6072 | 0.006 | 0.5416 | No | ||
| 39 | HOXA6 | 17150 | 6640 | 0.004 | 0.5112 | No | ||
| 40 | MSX1 | 16547 | 7667 | 0.001 | 0.4559 | No | ||
| 41 | UBD | 23239 | 8356 | -0.000 | 0.4187 | No | ||
| 42 | EGFL6 | 24006 | 8362 | -0.000 | 0.4185 | No | ||
| 43 | IL27 | 17636 | 8592 | -0.001 | 0.4062 | No | ||
| 44 | ASPA | 20354 | 8874 | -0.002 | 0.3911 | No | ||
| 45 | EIF4A2 | 4660 1679 1645 | 9078 | -0.002 | 0.3802 | No | ||
| 46 | BHLHB5 | 15629 | 9168 | -0.002 | 0.3755 | No | ||
| 47 | ATF3 | 13720 | 9228 | -0.002 | 0.3724 | No | ||
| 48 | PRDM16 | 12893 | 9383 | -0.003 | 0.3642 | No | ||
| 49 | NR3C2 | 18562 13798 | 9488 | -0.003 | 0.3586 | No | ||
| 50 | RFX4 | 7721 3410 | 9565 | -0.003 | 0.3547 | No | ||
| 51 | RIMS2 | 4370 | 10282 | -0.005 | 0.3162 | No | ||
| 52 | ARHGAP5 | 4412 8625 | 10422 | -0.005 | 0.3089 | No | ||
| 53 | PGM5 | 5928 | 11008 | -0.007 | 0.2776 | No | ||
| 54 | B2M | 72 14882 | 11141 | -0.008 | 0.2707 | No | ||
| 55 | NPR3 | 5188 | 11349 | -0.008 | 0.2598 | No | ||
| 56 | ASH1L | 5311 15546 15547 | 11855 | -0.010 | 0.2329 | No | ||
| 57 | ESR1 | 20097 4685 | 11982 | -0.011 | 0.2265 | No | ||
| 58 | SLC38A2 | 22149 2219 | 12064 | -0.011 | 0.2226 | No | ||
| 59 | PDGFC | 7048 | 12284 | -0.013 | 0.2112 | No | ||
| 60 | BNC2 | 15850 | 12671 | -0.015 | 0.1909 | No | ||
| 61 | GLRA1 | 9021 | 12674 | -0.015 | 0.1913 | No | ||
| 62 | TRPM3 | 23904 10357 5927 | 12743 | -0.016 | 0.1882 | No | ||
| 63 | TBX1 | 10039 5630 | 12979 | -0.017 | 0.1761 | No | ||
| 64 | MCM8 | 14834 | 13377 | -0.021 | 0.1554 | No | ||
| 65 | TGFB3 | 10161 | 13520 | -0.023 | 0.1486 | No | ||
| 66 | TMPRSS5 | 19463 | 13647 | -0.025 | 0.1426 | No | ||
| 67 | LIF | 956 667 20960 | 13887 | -0.028 | 0.1307 | No | ||
| 68 | TNFSF13B | 6291 10792 | 14105 | -0.032 | 0.1201 | No | ||
| 69 | IL15RA | 2899 2717 15118 2792 | 14202 | -0.034 | 0.1162 | No | ||
| 70 | TERF2IP | 12184 18463 7167 | 14334 | -0.038 | 0.1104 | No | ||
| 71 | PROX1 | 9623 | 14412 | -0.040 | 0.1076 | No | ||
| 72 | KCNE4 | 12196 | 14449 | -0.041 | 0.1071 | No | ||
| 73 | ROCK1 | 5386 | 14471 | -0.041 | 0.1074 | No | ||
| 74 | PIGR | 9561 | 14609 | -0.045 | 0.1016 | No | ||
| 75 | USF1 | 5832 10260 10261 | 14823 | -0.052 | 0.0919 | No | ||
| 76 | REL | 9716 | 15085 | -0.066 | 0.0802 | No | ||
| 77 | TAPBP | 5625 | 15377 | -0.084 | 0.0674 | No | ||
| 78 | BLK | 3205 21791 | 15497 | -0.094 | 0.0643 | No | ||
| 79 | DLG1 | 22793 1672 | 15564 | -0.101 | 0.0642 | No | ||
| 80 | NT5C3 | 17136 | 15807 | -0.129 | 0.0557 | No | ||
| 81 | USP5 | 17004 | 16525 | -0.273 | 0.0265 | No | ||
| 82 | E2F3 | 21500 8874 | 17131 | -0.456 | 0.0098 | No | ||
| 83 | KARS | 18738 | 17777 | -0.800 | 0.0029 | No | ||
| 84 | DAP3 | 7238 12271 1811 | 18178 | -1.211 | 0.0236 | No |