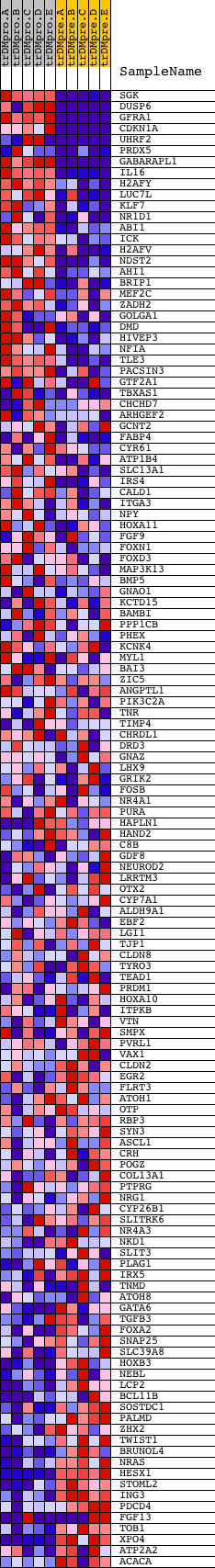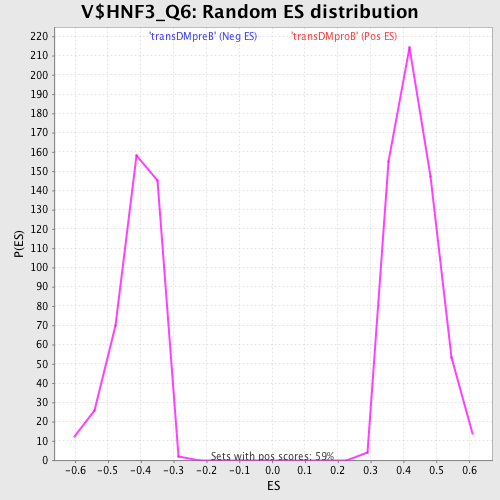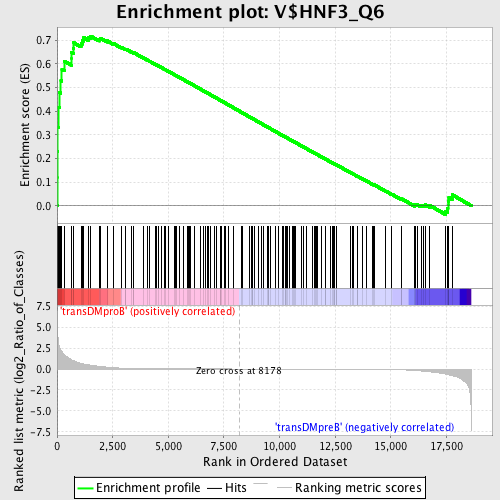
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$HNF3_Q6 |
| Enrichment Score (ES) | 0.7172756 |
| Normalized Enrichment Score (NES) | 1.659073 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.040469684 |
| FWER p-Value | 0.082 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SGK | 9809 3419 | 12 | 4.998 | 0.1209 | Yes | ||
| 2 | DUSP6 | 19891 3399 | 15 | 4.550 | 0.2314 | Yes | ||
| 3 | GFRA1 | 9015 | 28 | 4.130 | 0.3312 | Yes | ||
| 4 | CDKN1A | 4511 8729 | 49 | 3.556 | 0.4166 | Yes | ||
| 5 | UHRF2 | 3731 4262 8452 23887 | 99 | 2.667 | 0.4788 | Yes | ||
| 6 | PRDX5 | 7053 3732 | 172 | 2.242 | 0.5294 | Yes | ||
| 7 | GABARAPL1 | 17268 | 217 | 2.042 | 0.5767 | Yes | ||
| 8 | IL16 | 9171 | 329 | 1.661 | 0.6111 | Yes | ||
| 9 | H2AFY | 21447 | 637 | 1.098 | 0.6211 | Yes | ||
| 10 | LUC7L | 47 1520 | 642 | 1.092 | 0.6475 | Yes | ||
| 11 | KLF7 | 8205 | 734 | 0.981 | 0.6664 | Yes | ||
| 12 | NR1D1 | 4650 20258 949 | 749 | 0.966 | 0.6891 | Yes | ||
| 13 | ABI1 | 4300 | 1078 | 0.664 | 0.6875 | Yes | ||
| 14 | ICK | 7135 12146 | 1150 | 0.618 | 0.6987 | Yes | ||
| 15 | H2AFV | 1273 20531 | 1196 | 0.593 | 0.7107 | Yes | ||
| 16 | NDST2 | 21906 | 1422 | 0.493 | 0.7105 | Yes | ||
| 17 | AHI1 | 20077 | 1505 | 0.460 | 0.7173 | Yes | ||
| 18 | BRIP1 | 20311 | 1888 | 0.315 | 0.7043 | No | ||
| 19 | MEF2C | 3204 9378 | 1945 | 0.302 | 0.7086 | No | ||
| 20 | ZADH2 | 23501 | 2250 | 0.217 | 0.6974 | No | ||
| 21 | GOLGA1 | 14595 | 2532 | 0.156 | 0.6860 | No | ||
| 22 | DMD | 24295 2647 | 2886 | 0.102 | 0.6694 | No | ||
| 23 | HIVEP3 | 4973 | 3051 | 0.085 | 0.6626 | No | ||
| 24 | NFIA | 16172 5170 | 3093 | 0.082 | 0.6623 | No | ||
| 25 | TLE3 | 5767 19416 | 3343 | 0.060 | 0.6503 | No | ||
| 26 | PACSIN3 | 14948 8149 2776 2665 | 3442 | 0.054 | 0.6463 | No | ||
| 27 | GTF2A1 | 2064 8187 8188 13512 | 3452 | 0.053 | 0.6471 | No | ||
| 28 | TBXAS1 | 17480 1105 | 3879 | 0.033 | 0.6249 | No | ||
| 29 | CHCHD7 | 16280 | 4047 | 0.027 | 0.6165 | No | ||
| 30 | ARHGEF2 | 4987 | 4150 | 0.024 | 0.6116 | No | ||
| 31 | GCNT2 | 9168 3166 3243 21661 3270 | 4437 | 0.018 | 0.5966 | No | ||
| 32 | FABP4 | 8601 | 4444 | 0.018 | 0.5967 | No | ||
| 33 | CYR61 | 9159 15145 9311 5024 | 4566 | 0.016 | 0.5905 | No | ||
| 34 | ATP1B4 | 12586 | 4675 | 0.015 | 0.5850 | No | ||
| 35 | SLC13A1 | 17205 | 4808 | 0.013 | 0.5782 | No | ||
| 36 | IRS4 | 9183 4926 | 4849 | 0.013 | 0.5764 | No | ||
| 37 | CALD1 | 4273 8463 | 5005 | 0.012 | 0.5683 | No | ||
| 38 | ITGA3 | 20284 | 5278 | 0.010 | 0.5538 | No | ||
| 39 | NPY | 17449 | 5304 | 0.010 | 0.5527 | No | ||
| 40 | HOXA11 | 17146 | 5351 | 0.009 | 0.5504 | No | ||
| 41 | FGF9 | 8966 | 5506 | 0.009 | 0.5423 | No | ||
| 42 | FOXN1 | 20337 | 5684 | 0.008 | 0.5329 | No | ||
| 43 | FOXD3 | 9088 | 5849 | 0.007 | 0.5242 | No | ||
| 44 | MAP3K13 | 22814 | 5891 | 0.007 | 0.5221 | No | ||
| 45 | BMP5 | 19376 | 5941 | 0.007 | 0.5197 | No | ||
| 46 | GNAO1 | 4785 3829 | 6003 | 0.006 | 0.5165 | No | ||
| 47 | KCTD15 | 17864 | 6178 | 0.006 | 0.5072 | No | ||
| 48 | BAMBI | 23629 | 6458 | 0.005 | 0.4922 | No | ||
| 49 | PPP1CB | 5282 3529 3504 | 6599 | 0.004 | 0.4848 | No | ||
| 50 | PHEX | 24024 | 6685 | 0.004 | 0.4803 | No | ||
| 51 | KCNK4 | 3719 23803 | 6750 | 0.004 | 0.4769 | No | ||
| 52 | MYL1 | 4015 | 6797 | 0.004 | 0.4745 | No | ||
| 53 | BAI3 | 5580 | 6896 | 0.003 | 0.4693 | No | ||
| 54 | ZIC5 | 12268 | 7084 | 0.003 | 0.4592 | No | ||
| 55 | ANGPTL1 | 10195 8589 13064 | 7166 | 0.003 | 0.4549 | No | ||
| 56 | PIK3C2A | 17667 | 7330 | 0.002 | 0.4461 | No | ||
| 57 | TNR | 5787 | 7357 | 0.002 | 0.4448 | No | ||
| 58 | TIMP4 | 17036 | 7409 | 0.002 | 0.4421 | No | ||
| 59 | CHRDL1 | 8180 13504 | 7528 | 0.002 | 0.4357 | No | ||
| 60 | DRD3 | 22750 | 7579 | 0.002 | 0.4331 | No | ||
| 61 | GNAZ | 71 | 7699 | 0.001 | 0.4267 | No | ||
| 62 | LHX9 | 9276 | 7920 | 0.001 | 0.4148 | No | ||
| 63 | GRIK2 | 3318 | 8286 | -0.000 | 0.3950 | No | ||
| 64 | FOSB | 17945 | 8326 | -0.000 | 0.3929 | No | ||
| 65 | NR4A1 | 9099 | 8658 | -0.001 | 0.3751 | No | ||
| 66 | PURA | 9670 | 8758 | -0.001 | 0.3697 | No | ||
| 67 | HAPLN1 | 21592 | 8770 | -0.001 | 0.3692 | No | ||
| 68 | HAND2 | 18615 | 8794 | -0.001 | 0.3680 | No | ||
| 69 | C8B | 16163 | 8886 | -0.002 | 0.3631 | No | ||
| 70 | GDF8 | 9411 | 9055 | -0.002 | 0.3540 | No | ||
| 71 | NEUROD2 | 20262 | 9167 | -0.002 | 0.3481 | No | ||
| 72 | LRRTM3 | 19744 | 9277 | -0.003 | 0.3423 | No | ||
| 73 | OTX2 | 9519 | 9449 | -0.003 | 0.3331 | No | ||
| 74 | CYP7A1 | 15945 | 9480 | -0.003 | 0.3315 | No | ||
| 75 | ALDH9A1 | 14064 | 9491 | -0.003 | 0.3311 | No | ||
| 76 | EBF2 | 21975 | 9602 | -0.003 | 0.3252 | No | ||
| 77 | LGI1 | 23867 | 9798 | -0.004 | 0.3147 | No | ||
| 78 | TJP1 | 17803 | 9967 | -0.004 | 0.3057 | No | ||
| 79 | CLDN8 | 22549 1747 | 10152 | -0.005 | 0.2959 | No | ||
| 80 | TYRO3 | 5811 | 10170 | -0.005 | 0.2951 | No | ||
| 81 | TEAD1 | 18121 | 10267 | -0.005 | 0.2900 | No | ||
| 82 | PRDM1 | 19775 3337 | 10270 | -0.005 | 0.2900 | No | ||
| 83 | HOXA10 | 17145 989 | 10319 | -0.005 | 0.2875 | No | ||
| 84 | ITPKB | 3963 14028 | 10343 | -0.005 | 0.2864 | No | ||
| 85 | VTN | 20756 | 10466 | -0.005 | 0.2800 | No | ||
| 86 | SMPX | 24220 | 10578 | -0.006 | 0.2741 | No | ||
| 87 | PVRL1 | 3006 19482 12211 7192 | 10622 | -0.006 | 0.2719 | No | ||
| 88 | VAX1 | 23636 | 10658 | -0.006 | 0.2702 | No | ||
| 89 | CLDN2 | 4527 | 10728 | -0.006 | 0.2666 | No | ||
| 90 | EGR2 | 8886 | 10977 | -0.007 | 0.2533 | No | ||
| 91 | FLRT3 | 7730 12930 2673 | 11063 | -0.007 | 0.2489 | No | ||
| 92 | ATOH1 | 4419 | 11187 | -0.008 | 0.2425 | No | ||
| 93 | OTP | 21582 | 11477 | -0.009 | 0.2270 | No | ||
| 94 | RBP3 | 22047 | 11559 | -0.009 | 0.2229 | No | ||
| 95 | SYN3 | 19658 | 11602 | -0.009 | 0.2208 | No | ||
| 96 | ASCL1 | 19655 | 11670 | -0.010 | 0.2174 | No | ||
| 97 | CRH | 8784 | 11723 | -0.010 | 0.2149 | No | ||
| 98 | POGZ | 6031 1911 | 11885 | -0.011 | 0.2064 | No | ||
| 99 | COL13A1 | 8763 | 12057 | -0.011 | 0.1974 | No | ||
| 100 | PTPRG | 22097 3151 | 12062 | -0.011 | 0.1975 | No | ||
| 101 | NRG1 | 3885 | 12271 | -0.013 | 0.1866 | No | ||
| 102 | CYP26B1 | 17093 | 12357 | -0.013 | 0.1823 | No | ||
| 103 | SLITRK6 | 21724 | 12420 | -0.013 | 0.1792 | No | ||
| 104 | NR4A3 | 9473 16212 5183 | 12466 | -0.014 | 0.1771 | No | ||
| 105 | NKD1 | 8228 | 12481 | -0.014 | 0.1767 | No | ||
| 106 | SLIT3 | 20925 | 12553 | -0.014 | 0.1732 | No | ||
| 107 | PLAG1 | 7148 15949 12161 | 12574 | -0.014 | 0.1725 | No | ||
| 108 | IRX5 | 18527 | 13169 | -0.019 | 0.1408 | No | ||
| 109 | TNMD | 24260 | 13256 | -0.020 | 0.1366 | No | ||
| 110 | ATOH8 | 17120 | 13321 | -0.020 | 0.1337 | No | ||
| 111 | GATA6 | 96 | 13511 | -0.023 | 0.1240 | No | ||
| 112 | TGFB3 | 10161 | 13520 | -0.023 | 0.1241 | No | ||
| 113 | FOXA2 | 14405 2889 | 13734 | -0.026 | 0.1132 | No | ||
| 114 | SNAP25 | 5466 | 13903 | -0.029 | 0.1048 | No | ||
| 115 | SLC39A8 | 12547 1776 | 14177 | -0.034 | 0.0909 | No | ||
| 116 | HOXB3 | 20685 | 14240 | -0.035 | 0.0884 | No | ||
| 117 | NEBL | 14678 2691 | 14254 | -0.036 | 0.0886 | No | ||
| 118 | LCP2 | 4988 9268 | 14777 | -0.050 | 0.0615 | No | ||
| 119 | BCL11B | 7190 | 15046 | -0.064 | 0.0486 | No | ||
| 120 | SOSTDC1 | 21286 | 15468 | -0.092 | 0.0280 | No | ||
| 121 | PALMD | 15181 | 15496 | -0.094 | 0.0289 | No | ||
| 122 | ZHX2 | 11842 | 16078 | -0.171 | 0.0016 | No | ||
| 123 | TWIST1 | 21291 | 16092 | -0.175 | 0.0051 | No | ||
| 124 | BRUNOL4 | 2010 4229 | 16200 | -0.195 | 0.0041 | No | ||
| 125 | NRAS | 5191 | 16367 | -0.233 | 0.0008 | No | ||
| 126 | HESX1 | 22070 | 16470 | -0.259 | 0.0016 | No | ||
| 127 | STOML2 | 15902 | 16537 | -0.277 | 0.0047 | No | ||
| 128 | ING3 | 17514 1019 | 16747 | -0.331 | 0.0015 | No | ||
| 129 | PDCD4 | 5232 23816 | 17458 | -0.603 | -0.0223 | No | ||
| 130 | FGF13 | 4720 8963 2627 | 17551 | -0.656 | -0.0113 | No | ||
| 131 | TOB1 | 20703 | 17571 | -0.670 | 0.0040 | No | ||
| 132 | XPO4 | 7161 | 17574 | -0.673 | 0.0202 | No | ||
| 133 | ATP2A2 | 4421 3481 | 17610 | -0.694 | 0.0352 | No | ||
| 134 | ACACA | 309 4209 | 17758 | -0.787 | 0.0464 | No |