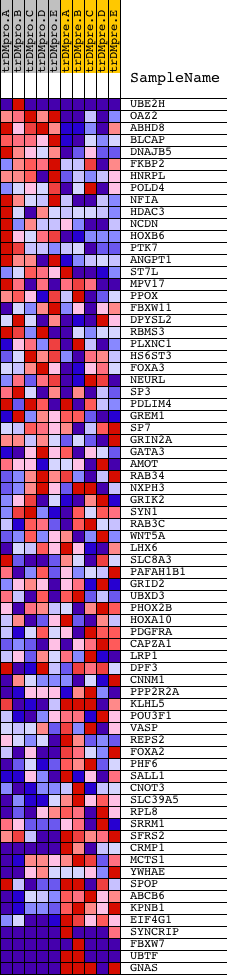
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
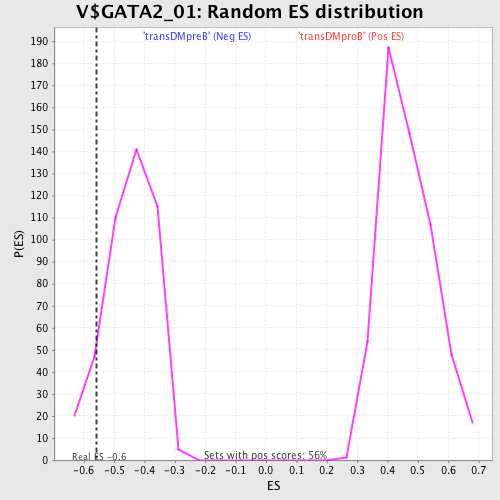
| GeneSet | V$GATA2_01 |
| Enrichment Score (ES) | -0.559108 |
| Normalized Enrichment Score (NES) | -1.2491018 |
| Nominal p-value | 0.09360731 |
| FDR q-value | 0.6963616 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2H | 5823 5822 | 402 | 1.542 | 0.0699 | No | ||
| 2 | OAZ2 | 9499 | 613 | 1.132 | 0.1259 | No | ||
| 3 | ABHD8 | 18847 | 1481 | 0.469 | 0.1070 | No | ||
| 4 | BLCAP | 14371 | 1583 | 0.421 | 0.1265 | No | ||
| 5 | DNAJB5 | 16236 | 1748 | 0.362 | 0.1392 | No | ||
| 6 | FKBP2 | 8972 | 2157 | 0.241 | 0.1316 | No | ||
| 7 | HNRPL | 18315 | 2620 | 0.142 | 0.1151 | No | ||
| 8 | POLD4 | 12822 | 2969 | 0.094 | 0.1019 | No | ||
| 9 | NFIA | 16172 5170 | 3093 | 0.082 | 0.1001 | No | ||
| 10 | HDAC3 | 4844 | 3253 | 0.067 | 0.0955 | No | ||
| 11 | NCDN | 15756 2487 6471 | 3623 | 0.043 | 0.0782 | No | ||
| 12 | HOXB6 | 20688 | 3942 | 0.031 | 0.0629 | No | ||
| 13 | PTK7 | 22959 | 3970 | 0.030 | 0.0632 | No | ||
| 14 | ANGPT1 | 8588 2260 22303 | 4133 | 0.025 | 0.0559 | No | ||
| 15 | ST7L | 15465 1904 | 4213 | 0.023 | 0.0530 | No | ||
| 16 | MPV17 | 9407 3505 3543 | 4251 | 0.022 | 0.0523 | No | ||
| 17 | PPOX | 13761 4104 | 4603 | 0.016 | 0.0344 | No | ||
| 18 | FBXW11 | 20926 | 4989 | 0.012 | 0.0143 | No | ||
| 19 | DPYSL2 | 3122 4561 8787 | 5140 | 0.011 | 0.0068 | No | ||
| 20 | RBMS3 | 18973 | 5155 | 0.011 | 0.0067 | No | ||
| 21 | PLXNC1 | 7056 | 5171 | 0.011 | 0.0065 | No | ||
| 22 | HS6ST3 | 21933 | 5305 | 0.010 | -0.0001 | No | ||
| 23 | FOXA3 | 17949 | 5450 | 0.009 | -0.0073 | No | ||
| 24 | NEURL | 5164 | 5997 | 0.006 | -0.0364 | No | ||
| 25 | SP3 | 5483 | 6119 | 0.006 | -0.0425 | No | ||
| 26 | PDLIM4 | 20455 | 6707 | 0.004 | -0.0740 | No | ||
| 27 | GREM1 | 14485 | 6777 | 0.004 | -0.0775 | No | ||
| 28 | SP7 | 22110 | 6928 | 0.003 | -0.0854 | No | ||
| 29 | GRIN2A | 22668 | 7235 | 0.002 | -0.1017 | No | ||
| 30 | GATA3 | 9004 | 7617 | 0.002 | -0.1222 | No | ||
| 31 | AMOT | 11341 24037 6587 | 7619 | 0.001 | -0.1221 | No | ||
| 32 | RAB34 | 20763 | 7714 | 0.001 | -0.1271 | No | ||
| 33 | NXPH3 | 20281 | 7895 | 0.001 | -0.1368 | No | ||
| 34 | GRIK2 | 3318 | 8286 | -0.000 | -0.1578 | No | ||
| 35 | SYN1 | 24175 5558 | 8289 | -0.000 | -0.1579 | No | ||
| 36 | RAB3C | 21351 | 8855 | -0.002 | -0.1883 | No | ||
| 37 | WNT5A | 22066 5880 | 8917 | -0.002 | -0.1915 | No | ||
| 38 | LHX6 | 9275 2914 | 9464 | -0.003 | -0.2207 | No | ||
| 39 | SLC8A3 | 2065 4293 | 9523 | -0.003 | -0.2237 | No | ||
| 40 | PAFAH1B1 | 1340 5220 9524 | 9572 | -0.003 | -0.2261 | No | ||
| 41 | GRID2 | 4803 | 9709 | -0.004 | -0.2332 | No | ||
| 42 | UBXD3 | 15701 | 9779 | -0.004 | -0.2367 | No | ||
| 43 | PHOX2B | 16524 | 9871 | -0.004 | -0.2414 | No | ||
| 44 | HOXA10 | 17145 989 | 10319 | -0.005 | -0.2652 | No | ||
| 45 | PDGFRA | 16824 | 10973 | -0.007 | -0.3000 | No | ||
| 46 | CAPZA1 | 8687 | 11078 | -0.007 | -0.3052 | No | ||
| 47 | LRP1 | 9284 | 11148 | -0.008 | -0.3084 | No | ||
| 48 | DPF3 | 7659 12853 | 11182 | -0.008 | -0.3097 | No | ||
| 49 | CNNM1 | 23846 | 11746 | -0.010 | -0.3395 | No | ||
| 50 | PPP2R2A | 3222 21774 | 12032 | -0.011 | -0.3542 | No | ||
| 51 | KLHL5 | 3631 7753 | 12519 | -0.014 | -0.3796 | No | ||
| 52 | POU3F1 | 16093 | 13065 | -0.018 | -0.4079 | No | ||
| 53 | VASP | 5847 | 13459 | -0.022 | -0.4278 | No | ||
| 54 | REPS2 | 24018 | 13554 | -0.023 | -0.4315 | No | ||
| 55 | FOXA2 | 14405 2889 | 13734 | -0.026 | -0.4396 | No | ||
| 56 | PHF6 | 2648 24340 | 13877 | -0.028 | -0.4456 | No | ||
| 57 | SALL1 | 18796 12207 | 13994 | -0.031 | -0.4500 | No | ||
| 58 | CNOT3 | 6112 10565 6113 | 14085 | -0.032 | -0.4529 | No | ||
| 59 | SLC39A5 | 3364 19596 | 14688 | -0.048 | -0.4826 | No | ||
| 60 | RPL8 | 22437 | 15856 | -0.136 | -0.5374 | Yes | ||
| 61 | SRRM1 | 6958 | 15872 | -0.139 | -0.5299 | Yes | ||
| 62 | SFRS2 | 9807 20136 | 15923 | -0.146 | -0.5240 | Yes | ||
| 63 | CRMP1 | 16860 | 16213 | -0.198 | -0.5278 | Yes | ||
| 64 | MCTS1 | 24351 | 16362 | -0.231 | -0.5220 | Yes | ||
| 65 | YWHAE | 20776 | 16449 | -0.254 | -0.5115 | Yes | ||
| 66 | SPOP | 5498 | 16637 | -0.305 | -0.5035 | Yes | ||
| 67 | ABCB6 | 13162 | 16884 | -0.371 | -0.4947 | Yes | ||
| 68 | KPNB1 | 20274 | 18079 | -1.073 | -0.4954 | Yes | ||
| 69 | EIF4G1 | 22818 | 18146 | -1.151 | -0.4305 | Yes | ||
| 70 | SYNCRIP | 3078 3035 3107 | 18203 | -1.259 | -0.3587 | Yes | ||
| 71 | FBXW7 | 1805 11928 | 18311 | -1.475 | -0.2768 | Yes | ||
| 72 | UBTF | 1313 10052 | 18449 | -1.857 | -0.1738 | Yes | ||
| 73 | GNAS | 9025 2963 2752 | 18570 | -3.076 | 0.0025 | Yes |