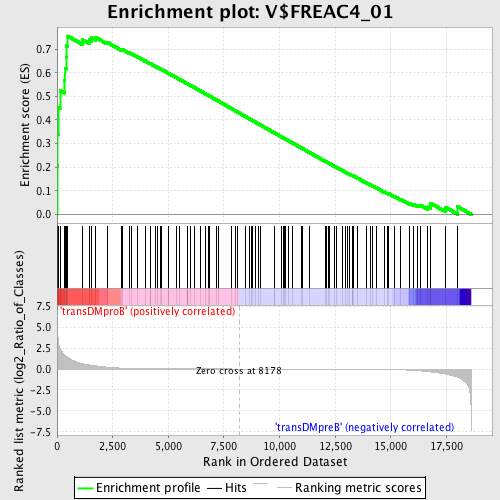
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | V$FREAC4_01 |
| Enrichment Score (ES) | 0.7550367 |
| Normalized Enrichment Score (NES) | 1.6791989 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07046526 |
| FWER p-Value | 0.049 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 17260 | 1 | 6.359 | 0.2055 | Yes | ||
| 2 | GFRA1 | 9015 | 28 | 4.130 | 0.3376 | Yes | ||
| 3 | CDKN1A | 4511 8729 | 49 | 3.556 | 0.4514 | Yes | ||
| 4 | LEF1 | 1860 15420 | 133 | 2.410 | 0.5248 | Yes | ||
| 5 | ETHE1 | 18347 | 334 | 1.654 | 0.5675 | Yes | ||
| 6 | TCF7L2 | 10048 5646 | 354 | 1.605 | 0.6184 | Yes | ||
| 7 | UBE2H | 5823 5822 | 402 | 1.542 | 0.6656 | Yes | ||
| 8 | DUSP1 | 23061 | 416 | 1.523 | 0.7142 | Yes | ||
| 9 | MBNL2 | 21932 | 484 | 1.376 | 0.7550 | Yes | ||
| 10 | PLS3 | 8332 | 1145 | 0.620 | 0.7394 | No | ||
| 11 | ZFX | 5984 | 1438 | 0.487 | 0.7394 | No | ||
| 12 | CTCF | 18490 | 1551 | 0.438 | 0.7475 | No | ||
| 13 | KBTBD2 | 17137 | 1731 | 0.368 | 0.7498 | No | ||
| 14 | ZADH2 | 23501 | 2250 | 0.217 | 0.7288 | No | ||
| 15 | ULK1 | 16436 | 2912 | 0.099 | 0.6963 | No | ||
| 16 | TRPS1 | 8195 | 2925 | 0.098 | 0.6988 | No | ||
| 17 | HEPH | 24288 2623 2635 2568 2652 | 3243 | 0.068 | 0.6839 | No | ||
| 18 | TLE3 | 5767 19416 | 3343 | 0.060 | 0.6805 | No | ||
| 19 | NCDN | 15756 2487 6471 | 3623 | 0.043 | 0.6668 | No | ||
| 20 | EYA1 | 4695 4061 | 3955 | 0.030 | 0.6500 | No | ||
| 21 | OTOP3 | 20599 | 4178 | 0.024 | 0.6387 | No | ||
| 22 | PELI2 | 8217 | 4180 | 0.024 | 0.6395 | No | ||
| 23 | FOXB1 | 12254 19069 | 4427 | 0.018 | 0.6268 | No | ||
| 24 | TCF1 | 16416 | 4523 | 0.017 | 0.6222 | No | ||
| 25 | MSC | 13998 | 4626 | 0.015 | 0.6172 | No | ||
| 26 | CAMK1D | 5977 2948 | 4690 | 0.015 | 0.6143 | No | ||
| 27 | CALD1 | 4273 8463 | 5005 | 0.012 | 0.5977 | No | ||
| 28 | OTX1 | 20517 | 5019 | 0.012 | 0.5974 | No | ||
| 29 | HOXA11 | 17146 | 5351 | 0.009 | 0.5798 | No | ||
| 30 | FGF9 | 8966 | 5506 | 0.009 | 0.5718 | No | ||
| 31 | FOXD3 | 9088 | 5849 | 0.007 | 0.5535 | No | ||
| 32 | VIT | 23155 | 5867 | 0.007 | 0.5528 | No | ||
| 33 | ROR1 | 6472 | 5871 | 0.007 | 0.5529 | No | ||
| 34 | SYT10 | 22157 | 6001 | 0.006 | 0.5461 | No | ||
| 35 | SHOX2 | 1802 5432 | 6163 | 0.006 | 0.5376 | No | ||
| 36 | LOX | 23438 | 6462 | 0.005 | 0.5217 | No | ||
| 37 | PHEX | 24024 | 6685 | 0.004 | 0.5098 | No | ||
| 38 | PNRC1 | 15925 | 6825 | 0.004 | 0.5024 | No | ||
| 39 | GABRR1 | 16250 | 6846 | 0.003 | 0.5014 | No | ||
| 40 | PITX2 | 15424 1878 | 7147 | 0.003 | 0.4853 | No | ||
| 41 | TLX1 | 23839 | 7241 | 0.002 | 0.4804 | No | ||
| 42 | HOXD9 | 14982 | 7822 | 0.001 | 0.4491 | No | ||
| 43 | SLC24A2 | 8020 13326 8021 | 8019 | 0.000 | 0.4385 | No | ||
| 44 | NNAT | 2764 5180 | 8129 | 0.000 | 0.4327 | No | ||
| 45 | ODZ1 | 24168 | 8484 | -0.001 | 0.4136 | No | ||
| 46 | NR4A1 | 9099 | 8658 | -0.001 | 0.4043 | No | ||
| 47 | PURA | 9670 | 8758 | -0.001 | 0.3990 | No | ||
| 48 | PER2 | 3940 13881 | 8769 | -0.001 | 0.3985 | No | ||
| 49 | MYH2 | 20838 | 8935 | -0.002 | 0.3896 | No | ||
| 50 | SPRY4 | 23448 | 9072 | -0.002 | 0.3823 | No | ||
| 51 | NEDD4 | 19384 3101 | 9119 | -0.002 | 0.3799 | No | ||
| 52 | IBSP | 16775 | 9762 | -0.004 | 0.3454 | No | ||
| 53 | UBXD3 | 15701 | 9779 | -0.004 | 0.3446 | No | ||
| 54 | SEMA5B | 9801 22777 | 10084 | -0.004 | 0.3284 | No | ||
| 55 | HOXC6 | 22339 2302 | 10191 | -0.005 | 0.3228 | No | ||
| 56 | CD109 | 6174 10656 19369 | 10221 | -0.005 | 0.3214 | No | ||
| 57 | PRDM1 | 19775 3337 | 10270 | -0.005 | 0.3190 | No | ||
| 58 | OSR2 | 22492 | 10389 | -0.005 | 0.3128 | No | ||
| 59 | PLUNC | 9593 | 10576 | -0.006 | 0.3029 | No | ||
| 60 | EVX1 | 17442 | 10982 | -0.007 | 0.2813 | No | ||
| 61 | SLC34A3 | 14664 | 11050 | -0.007 | 0.2779 | No | ||
| 62 | IER5 | 13802 | 11344 | -0.008 | 0.2624 | No | ||
| 63 | CITED2 | 5118 14477 | 12067 | -0.011 | 0.2237 | No | ||
| 64 | PTHR1 | 5318 18985 | 12112 | -0.012 | 0.2217 | No | ||
| 65 | RARG | 22113 | 12219 | -0.012 | 0.2164 | No | ||
| 66 | CLDN18 | 19028 | 12242 | -0.012 | 0.2156 | No | ||
| 67 | ADRB1 | 4357 | 12463 | -0.014 | 0.2042 | No | ||
| 68 | COLEC12 | 23624 8946 | 12538 | -0.014 | 0.2007 | No | ||
| 69 | BTBD3 | 10457 2975 6007 6006 | 12821 | -0.016 | 0.1860 | No | ||
| 70 | PDCD1LG2 | 23892 | 12975 | -0.017 | 0.1783 | No | ||
| 71 | HR | 3622 9119 | 13059 | -0.018 | 0.1744 | No | ||
| 72 | KLF12 | 4960 2637 21732 9228 | 13156 | -0.019 | 0.1698 | No | ||
| 73 | TNMD | 24260 | 13256 | -0.020 | 0.1651 | No | ||
| 74 | ERG | 1686 8915 | 13281 | -0.020 | 0.1644 | No | ||
| 75 | POU3F3 | 14261 | 13339 | -0.021 | 0.1620 | No | ||
| 76 | TGFB3 | 10161 | 13520 | -0.023 | 0.1530 | No | ||
| 77 | TECTA | 19159 | 13884 | -0.028 | 0.1344 | No | ||
| 78 | AP1G2 | 2677 21827 | 14069 | -0.032 | 0.1255 | No | ||
| 79 | ALDH1A2 | 19386 | 14170 | -0.034 | 0.1211 | No | ||
| 80 | FOXP2 | 4312 17523 8523 | 14370 | -0.039 | 0.1116 | No | ||
| 81 | IGF1 | 3352 9156 3409 | 14702 | -0.048 | 0.0953 | No | ||
| 82 | TBX4 | 5633 | 14830 | -0.053 | 0.0902 | No | ||
| 83 | FERD3L | 21292 | 14904 | -0.056 | 0.0880 | No | ||
| 84 | MCC | 552 | 15167 | -0.071 | 0.0762 | No | ||
| 85 | HOXC4 | 4864 | 15454 | -0.090 | 0.0637 | No | ||
| 86 | SIN3A | 5442 | 15834 | -0.133 | 0.0475 | No | ||
| 87 | HSPG2 | 4884 16023 | 16031 | -0.164 | 0.0422 | No | ||
| 88 | PHTF2 | 16599 | 16203 | -0.195 | 0.0393 | No | ||
| 89 | TMOD3 | 6945 | 16334 | -0.225 | 0.0395 | No | ||
| 90 | SLC31A1 | 2447 16195 | 16667 | -0.312 | 0.0317 | No | ||
| 91 | TAF5L | 18712 | 16779 | -0.340 | 0.0367 | No | ||
| 92 | PDE7A | 9544 1788 | 16789 | -0.343 | 0.0473 | No | ||
| 93 | PUM2 | 2071 8161 | 17457 | -0.602 | 0.0308 | No | ||
| 94 | LMO4 | 15151 | 18004 | -0.982 | 0.0330 | No |