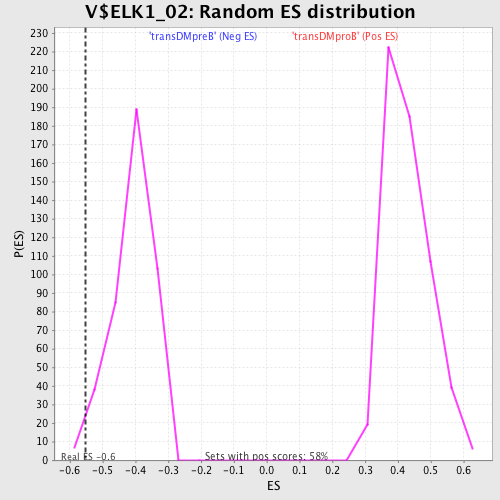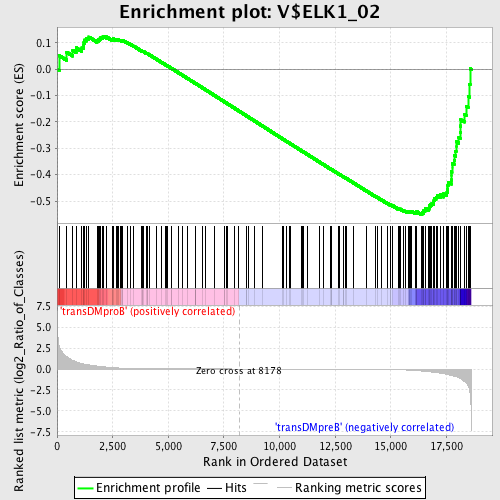
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | V$ELK1_02 |
| Enrichment Score (ES) | -0.55100006 |
| Normalized Enrichment Score (NES) | -1.3531905 |
| Nominal p-value | 0.016587678 |
| FDR q-value | 0.99728334 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITM2C | 14204 | 93 | 2.732 | 0.0521 | No | ||
| 2 | GTF3C1 | 10607 17646 | 435 | 1.488 | 0.0647 | No | ||
| 3 | PRKACB | 15140 | 705 | 1.014 | 0.0713 | No | ||
| 4 | KRTCAP2 | 15541 | 855 | 0.871 | 0.0814 | No | ||
| 5 | RAP2C | 24156 | 1116 | 0.642 | 0.0808 | No | ||
| 6 | TBCC | 23215 | 1182 | 0.601 | 0.0898 | No | ||
| 7 | RAB11B | 9676 | 1204 | 0.591 | 0.1010 | No | ||
| 8 | CGGBP1 | 22724 | 1241 | 0.574 | 0.1111 | No | ||
| 9 | DPP8 | 19408 | 1316 | 0.540 | 0.1183 | No | ||
| 10 | SEC24C | 22086 | 1431 | 0.490 | 0.1224 | No | ||
| 11 | FBXW9 | 18540 | 1793 | 0.346 | 0.1101 | No | ||
| 12 | UQCRH | 12378 | 1863 | 0.323 | 0.1131 | No | ||
| 13 | STK4 | 14743 | 1926 | 0.308 | 0.1162 | No | ||
| 14 | NEDD1 | 19643 | 1964 | 0.297 | 0.1204 | No | ||
| 15 | HMG20A | 19436 | 2034 | 0.275 | 0.1224 | No | ||
| 16 | NR1H2 | 17846 | 2075 | 0.263 | 0.1257 | No | ||
| 17 | AP1B1 | 8599 | 2216 | 0.225 | 0.1229 | No | ||
| 18 | PURB | 5335 | 2487 | 0.163 | 0.1116 | No | ||
| 19 | PCYT1A | 4572 | 2497 | 0.161 | 0.1145 | No | ||
| 20 | GOLGA1 | 14595 | 2532 | 0.156 | 0.1160 | No | ||
| 21 | ARHGAP1 | 6001 10448 | 2662 | 0.136 | 0.1118 | No | ||
| 22 | TBC1D17 | 6121 | 2692 | 0.131 | 0.1130 | No | ||
| 23 | FBXO36 | 14205 | 2737 | 0.123 | 0.1132 | No | ||
| 24 | MYST2 | 20283 | 2842 | 0.108 | 0.1098 | No | ||
| 25 | RPL19 | 9736 | 2905 | 0.100 | 0.1085 | No | ||
| 26 | TBX6 | 1993 18075 | 2940 | 0.096 | 0.1087 | No | ||
| 27 | ELAVL1 | 4888 | 2945 | 0.096 | 0.1105 | No | ||
| 28 | RPS18 | 1529 5397 1603 | 3143 | 0.077 | 0.1014 | No | ||
| 29 | VPS52 | 23287 1622 | 3290 | 0.064 | 0.0948 | No | ||
| 30 | NF1 | 5165 | 3443 | 0.054 | 0.0877 | No | ||
| 31 | GTF3C2 | 7749 | 3781 | 0.036 | 0.0702 | No | ||
| 32 | SIRT6 | 19668 3447 | 3816 | 0.036 | 0.0691 | No | ||
| 33 | RAPH1 | 19110 | 3845 | 0.034 | 0.0683 | No | ||
| 34 | BMP2K | 3657 16786 | 3876 | 0.033 | 0.0674 | No | ||
| 35 | COG3 | 21751 | 4022 | 0.028 | 0.0602 | No | ||
| 36 | ZDHHC5 | 14547 | 4058 | 0.027 | 0.0588 | No | ||
| 37 | BAT3 | 5896 | 4074 | 0.027 | 0.0586 | No | ||
| 38 | ZNF22 | 12497 7417 | 4170 | 0.024 | 0.0539 | No | ||
| 39 | CRB3 | 23176 | 4481 | 0.018 | 0.0375 | No | ||
| 40 | UBR1 | 5828 | 4693 | 0.015 | 0.0264 | No | ||
| 41 | CALU | 1124 1149 8683 1162 4472 | 4874 | 0.013 | 0.0169 | No | ||
| 42 | TRO | 7071 | 4900 | 0.013 | 0.0158 | No | ||
| 43 | SHC1 | 9813 9812 5430 | 4950 | 0.012 | 0.0134 | No | ||
| 44 | TRIM46 | 1779 15281 | 5130 | 0.011 | 0.0040 | No | ||
| 45 | CRK | 4559 1249 | 5134 | 0.011 | 0.0040 | No | ||
| 46 | BCL2L1 | 4440 2930 8652 | 5474 | 0.009 | -0.0142 | No | ||
| 47 | PCQAP | 1661 13581 | 5644 | 0.008 | -0.0231 | No | ||
| 48 | SYT5 | 17986 | 5860 | 0.007 | -0.0346 | No | ||
| 49 | ZBTB11 | 11309 | 6198 | 0.006 | -0.0528 | No | ||
| 50 | ITGB1BP1 | 9189 | 6207 | 0.006 | -0.0531 | No | ||
| 51 | ACP2 | 4318 | 6229 | 0.005 | -0.0541 | No | ||
| 52 | NUDT12 | 7518 | 6527 | 0.004 | -0.0701 | No | ||
| 53 | PAPD4 | 8299 8300 4146 | 6673 | 0.004 | -0.0779 | No | ||
| 54 | PPIE | 15766 | 7057 | 0.003 | -0.0985 | No | ||
| 55 | LSM5 | 7286 | 7525 | 0.002 | -0.1238 | No | ||
| 56 | FOXH1 | 22238 | 7541 | 0.002 | -0.1246 | No | ||
| 57 | GGTL3 | 14381 | 7605 | 0.002 | -0.1280 | No | ||
| 58 | STX12 | 4140 | 7680 | 0.001 | -0.1319 | No | ||
| 59 | FBXO3 | 2662 14934 | 7952 | 0.001 | -0.1466 | No | ||
| 60 | FBXO22 | 19442 | 8139 | 0.000 | -0.1567 | No | ||
| 61 | CLDN7 | 20817 | 8504 | -0.001 | -0.1764 | No | ||
| 62 | SON | 5473 1657 1684 | 8508 | -0.001 | -0.1765 | No | ||
| 63 | SRP19 | 2004 12341 | 8620 | -0.001 | -0.1825 | No | ||
| 64 | ACTR2 | 20523 | 8867 | -0.002 | -0.1958 | No | ||
| 65 | NFYA | 5172 | 9209 | -0.002 | -0.2142 | No | ||
| 66 | INSIG2 | 3944 4124 4073 | 10110 | -0.005 | -0.2628 | No | ||
| 67 | COX8A | 8777 | 10169 | -0.005 | -0.2659 | No | ||
| 68 | NDUFS1 | 13940 | 10312 | -0.005 | -0.2735 | No | ||
| 69 | FMR1 | 24321 | 10437 | -0.005 | -0.2801 | No | ||
| 70 | NAT5 | 18601 7496 12597 | 10468 | -0.006 | -0.2816 | No | ||
| 71 | MAPK8IP3 | 11402 23080 | 11006 | -0.007 | -0.3105 | No | ||
| 72 | RBM22 | 23542 | 11039 | -0.007 | -0.3121 | No | ||
| 73 | CPSF3 | 7038 | 11073 | -0.007 | -0.3137 | No | ||
| 74 | IMMT | 367 8029 | 11238 | -0.008 | -0.3224 | No | ||
| 75 | LYPLA2 | 15708 | 11777 | -0.010 | -0.3513 | No | ||
| 76 | GTF2A2 | 10654 | 11981 | -0.011 | -0.3621 | No | ||
| 77 | EN1 | 387 14155 | 12307 | -0.013 | -0.3794 | No | ||
| 78 | DDOST | 16021 | 12336 | -0.013 | -0.3807 | No | ||
| 79 | CDC45L | 22642 1752 | 12341 | -0.013 | -0.3806 | No | ||
| 80 | DDX5 | 4606 8843 4607 | 12656 | -0.015 | -0.3973 | No | ||
| 81 | PEX11A | 1056 5241 | 12675 | -0.015 | -0.3980 | No | ||
| 82 | MTX2 | 14980 | 12852 | -0.016 | -0.4071 | No | ||
| 83 | CLDN5 | 22826 | 12876 | -0.017 | -0.4080 | No | ||
| 84 | OGG1 | 17331 | 12951 | -0.017 | -0.4117 | No | ||
| 85 | LEPRE1 | 2384 12122 | 13008 | -0.018 | -0.4144 | No | ||
| 86 | DNAJA2 | 12133 18806 | 13334 | -0.020 | -0.4315 | No | ||
| 87 | GABARAPL2 | 8211 13552 | 13899 | -0.029 | -0.4615 | No | ||
| 88 | TRIM44 | 8164 13487 | 14288 | -0.036 | -0.4817 | No | ||
| 89 | DIABLO | 16375 | 14401 | -0.039 | -0.4870 | No | ||
| 90 | UFD1L | 22825 | 14588 | -0.045 | -0.4961 | No | ||
| 91 | ZNF23 | 6149 | 14848 | -0.054 | -0.5090 | No | ||
| 92 | TCF7 | 1467 20466 | 14977 | -0.060 | -0.5147 | No | ||
| 93 | RPL37 | 12502 7421 22521 | 14998 | -0.061 | -0.5145 | No | ||
| 94 | SAMD8 | 22082 | 15095 | -0.067 | -0.5183 | No | ||
| 95 | IL2RG | 24096 | 15322 | -0.080 | -0.5289 | No | ||
| 96 | SUPT5H | 9939 | 15388 | -0.085 | -0.5306 | No | ||
| 97 | EPN1 | 18401 | 15413 | -0.087 | -0.5301 | No | ||
| 98 | PSMC6 | 7379 | 15578 | -0.102 | -0.5368 | No | ||
| 99 | PHKB | 18537 3873 | 15648 | -0.110 | -0.5383 | No | ||
| 100 | TSTA3 | 22253 | 15775 | -0.125 | -0.5425 | No | ||
| 101 | TRAF7 | 10325 | 15823 | -0.132 | -0.5423 | No | ||
| 102 | UBE2L3 | 5818 22647 | 15824 | -0.132 | -0.5395 | No | ||
| 103 | HMGA1 | 23323 | 15876 | -0.140 | -0.5394 | No | ||
| 104 | NKIRAS2 | 12979 | 15942 | -0.148 | -0.5398 | No | ||
| 105 | SMUG1 | 22109 | 16111 | -0.179 | -0.5452 | No | ||
| 106 | ARCN1 | 19143 | 16159 | -0.188 | -0.5438 | No | ||
| 107 | AP1M1 | 18571 | 16161 | -0.188 | -0.5399 | No | ||
| 108 | NRAS | 5191 | 16367 | -0.233 | -0.5461 | Yes | ||
| 109 | LMAN2 | 21454 | 16425 | -0.249 | -0.5440 | Yes | ||
| 110 | UBE2N | 8216 19898 | 16465 | -0.258 | -0.5407 | Yes | ||
| 111 | AKT1S1 | 12557 | 16483 | -0.264 | -0.5361 | Yes | ||
| 112 | STOML2 | 15902 | 16537 | -0.277 | -0.5332 | Yes | ||
| 113 | HSPC171 | 18494 | 16558 | -0.283 | -0.5284 | Yes | ||
| 114 | PSMC1 | 9635 | 16701 | -0.319 | -0.5294 | Yes | ||
| 115 | MTPN | 17183 | 16718 | -0.324 | -0.5235 | Yes | ||
| 116 | DHX30 | 18989 | 16737 | -0.329 | -0.5176 | Yes | ||
| 117 | HAT1 | 14990 | 16784 | -0.341 | -0.5129 | Yes | ||
| 118 | SNRPB | 9842 5469 2736 | 16845 | -0.360 | -0.5087 | Yes | ||
| 119 | KLHDC3 | 22956 | 16919 | -0.383 | -0.5046 | Yes | ||
| 120 | PSMC2 | 16909 | 16937 | -0.388 | -0.4974 | Yes | ||
| 121 | UBADC1 | 2962 13608 | 16966 | -0.396 | -0.4907 | Yes | ||
| 122 | MIR16 | 17663 1354 | 17032 | -0.423 | -0.4854 | Yes | ||
| 123 | TOMM40 | 6997 | 17078 | -0.436 | -0.4787 | Yes | ||
| 124 | PSMD12 | 20621 | 17224 | -0.490 | -0.4763 | Yes | ||
| 125 | SLC30A7 | 15186 | 17381 | -0.570 | -0.4728 | Yes | ||
| 126 | TIMM8B | 11391 | 17521 | -0.640 | -0.4670 | Yes | ||
| 127 | POLL | 23658 3688 | 17537 | -0.649 | -0.4542 | Yes | ||
| 128 | SMS | 2573 24023 | 17563 | -0.666 | -0.4417 | Yes | ||
| 129 | CHERP | 11367 | 17577 | -0.674 | -0.4283 | Yes | ||
| 130 | PRKAG2 | 24 | 17711 | -0.746 | -0.4199 | Yes | ||
| 131 | TRMT1 | 2307 12985 | 17724 | -0.757 | -0.4047 | Yes | ||
| 132 | DNAJC7 | 20227 | 17727 | -0.759 | -0.3890 | Yes | ||
| 133 | ATP5A1 | 23505 | 17750 | -0.782 | -0.3738 | Yes | ||
| 134 | FANCD2 | 17326 464 | 17751 | -0.782 | -0.3575 | Yes | ||
| 135 | MARK3 | 9369 | 17845 | -0.854 | -0.3447 | Yes | ||
| 136 | TUFM | 10608 | 17869 | -0.868 | -0.3278 | Yes | ||
| 137 | EEF1B2 | 4131 12063 | 17910 | -0.902 | -0.3111 | Yes | ||
| 138 | SDHD | 19125 | 17934 | -0.918 | -0.2932 | Yes | ||
| 139 | RFC4 | 1735 22627 | 17938 | -0.919 | -0.2741 | Yes | ||
| 140 | IPO4 | 7983 | 18057 | -1.045 | -0.2587 | Yes | ||
| 141 | MTBP | 22473 8374 | 18117 | -1.129 | -0.2383 | Yes | ||
| 142 | EXTL2 | 15440 | 18130 | -1.139 | -0.2151 | Yes | ||
| 143 | EIF4G1 | 22818 | 18146 | -1.151 | -0.1919 | Yes | ||
| 144 | EIF5A | 11345 20379 6590 | 18300 | -1.448 | -0.1699 | Yes | ||
| 145 | GART | 22543 1754 | 18389 | -1.632 | -0.1406 | Yes | ||
| 146 | MAPKAP1 | 15030 5993 | 18475 | -2.019 | -0.1030 | Yes | ||
| 147 | MRPL3 | 3116 19335 | 18528 | -2.380 | -0.0561 | Yes | ||
| 148 | MRPL13 | 2249 22294 | 18565 | -2.909 | 0.0028 | Yes |