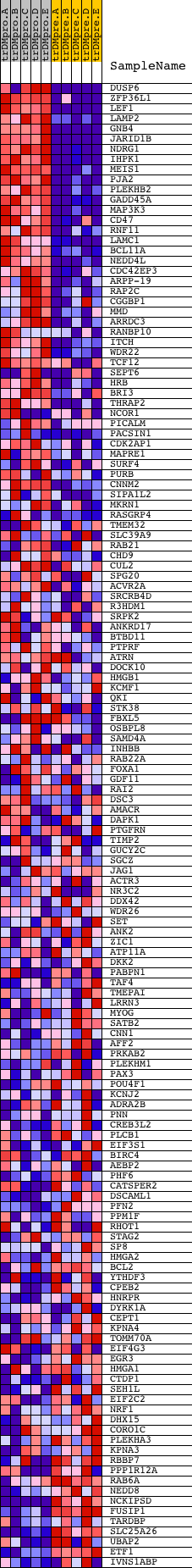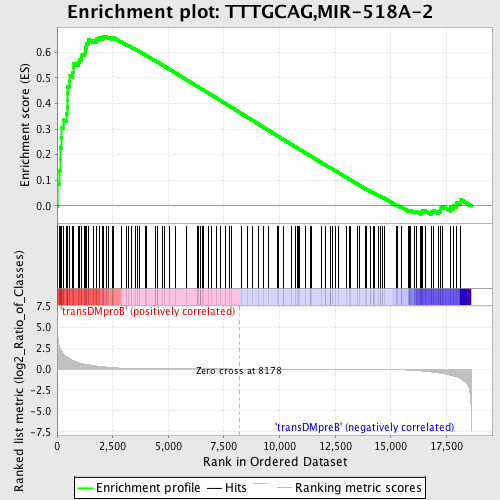
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | TTTGCAG,MIR-518A-2 |
| Enrichment Score (ES) | 0.6625491 |
| Normalized Enrichment Score (NES) | 1.5423148 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07313076 |
| FWER p-Value | 0.566 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DUSP6 | 19891 3399 | 15 | 4.550 | 0.0878 | Yes | ||
| 2 | ZFP36L1 | 4453 4454 | 94 | 2.717 | 0.1365 | Yes | ||
| 3 | LEF1 | 1860 15420 | 133 | 2.410 | 0.1814 | Yes | ||
| 4 | LAMP2 | 9267 2653 | 136 | 2.390 | 0.2278 | Yes | ||
| 5 | GNB4 | 4787 9028 | 181 | 2.213 | 0.2686 | Yes | ||
| 6 | JARID1B | 14123 | 216 | 2.046 | 0.3066 | Yes | ||
| 7 | NDRG1 | 22276 | 305 | 1.727 | 0.3355 | Yes | ||
| 8 | IHPK1 | 11336 | 411 | 1.533 | 0.3596 | Yes | ||
| 9 | MEIS1 | 20524 | 446 | 1.464 | 0.3863 | Yes | ||
| 10 | PJA2 | 5904 | 469 | 1.407 | 0.4125 | Yes | ||
| 11 | PLEKHB2 | 10390 | 472 | 1.398 | 0.4396 | Yes | ||
| 12 | GADD45A | 17129 | 481 | 1.383 | 0.4661 | Yes | ||
| 13 | MAP3K3 | 20626 | 561 | 1.226 | 0.4857 | Yes | ||
| 14 | CD47 | 4933 | 578 | 1.200 | 0.5082 | Yes | ||
| 15 | RNF11 | 11383 2509 | 704 | 1.015 | 0.5212 | Yes | ||
| 16 | LAMC1 | 13805 | 718 | 1.002 | 0.5400 | Yes | ||
| 17 | BCL11A | 4691 | 748 | 0.967 | 0.5573 | Yes | ||
| 18 | NEDD4L | 8192 | 951 | 0.776 | 0.5615 | Yes | ||
| 19 | CDC42EP3 | 6436 | 1007 | 0.722 | 0.5726 | Yes | ||
| 20 | ARPP-19 | 19379 | 1099 | 0.654 | 0.5804 | Yes | ||
| 21 | RAP2C | 24156 | 1116 | 0.642 | 0.5920 | Yes | ||
| 22 | CGGBP1 | 22724 | 1241 | 0.574 | 0.5965 | Yes | ||
| 23 | MMD | 20706 | 1261 | 0.564 | 0.6064 | Yes | ||
| 24 | ARRDC3 | 4191 | 1270 | 0.561 | 0.6169 | Yes | ||
| 25 | RANBP10 | 18764 | 1327 | 0.533 | 0.6243 | Yes | ||
| 26 | ITCH | 4928 9185 | 1329 | 0.533 | 0.6346 | Yes | ||
| 27 | WDR22 | 6803 | 1395 | 0.505 | 0.6409 | Yes | ||
| 28 | TCF12 | 10044 3125 | 1417 | 0.495 | 0.6494 | Yes | ||
| 29 | SEPT6 | 2591 7132 | 1639 | 0.401 | 0.6453 | Yes | ||
| 30 | HRB | 14208 4066 | 1758 | 0.361 | 0.6459 | Yes | ||
| 31 | BRI3 | 15101 | 1773 | 0.355 | 0.6521 | Yes | ||
| 32 | THRAP2 | 8009 | 1883 | 0.318 | 0.6523 | Yes | ||
| 33 | NCOR1 | 5402 | 1918 | 0.311 | 0.6566 | Yes | ||
| 34 | PICALM | 18191 | 2061 | 0.266 | 0.6541 | Yes | ||
| 35 | PACSIN1 | 6257 23322 10744 | 2076 | 0.263 | 0.6584 | Yes | ||
| 36 | CDK2AP1 | 8858 | 2094 | 0.259 | 0.6625 | Yes | ||
| 37 | MAPRE1 | 4652 | 2197 | 0.230 | 0.6615 | No | ||
| 38 | SURF4 | 14646 | 2326 | 0.198 | 0.6584 | No | ||
| 39 | PURB | 5335 | 2487 | 0.163 | 0.6529 | No | ||
| 40 | CNNM2 | 8251 | 2488 | 0.163 | 0.6561 | No | ||
| 41 | SIPA1L2 | 10863 | 2514 | 0.158 | 0.6578 | No | ||
| 42 | MKRN1 | 17178 | 2880 | 0.102 | 0.6401 | No | ||
| 43 | RASGRP4 | 6119 | 3132 | 0.078 | 0.6280 | No | ||
| 44 | TMEM32 | 24149 | 3208 | 0.071 | 0.6253 | No | ||
| 45 | SLC39A9 | 11597 | 3328 | 0.061 | 0.6201 | No | ||
| 46 | RAB21 | 5696 | 3543 | 0.047 | 0.6094 | No | ||
| 47 | CHD9 | 6784 11553 18531 | 3615 | 0.043 | 0.6064 | No | ||
| 48 | CUL2 | 23630 | 3714 | 0.039 | 0.6019 | No | ||
| 49 | SPG20 | 6024 | 3993 | 0.029 | 0.5874 | No | ||
| 50 | ACVR2A | 8542 | 4035 | 0.028 | 0.5857 | No | ||
| 51 | SRCRB4D | 8459 | 4405 | 0.019 | 0.5661 | No | ||
| 52 | R3HDM1 | 4002 974 | 4419 | 0.019 | 0.5658 | No | ||
| 53 | SRPK2 | 5513 | 4494 | 0.017 | 0.5621 | No | ||
| 54 | ANKRD17 | 13496 3474 8170 | 4499 | 0.017 | 0.5622 | No | ||
| 55 | BTBD11 | 13147 | 4736 | 0.014 | 0.5497 | No | ||
| 56 | PTPRF | 5331 2545 | 4823 | 0.013 | 0.5453 | No | ||
| 57 | ATRN | 4432 2964 | 5038 | 0.011 | 0.5339 | No | ||
| 58 | DOCK10 | 4004 13904 9970 | 5303 | 0.010 | 0.5198 | No | ||
| 59 | HMGB1 | 9094 4855 | 5826 | 0.007 | 0.4917 | No | ||
| 60 | KCMF1 | 17112 | 6297 | 0.005 | 0.4664 | No | ||
| 61 | QKI | 23128 5340 | 6318 | 0.005 | 0.4654 | No | ||
| 62 | STK38 | 23045 | 6366 | 0.005 | 0.4629 | No | ||
| 63 | FBXL5 | 6330 10828 16540 | 6436 | 0.005 | 0.4593 | No | ||
| 64 | OSBPL8 | 6198 | 6437 | 0.005 | 0.4594 | No | ||
| 65 | SAMD4A | 13211 | 6450 | 0.005 | 0.4588 | No | ||
| 66 | INHBB | 13858 | 6549 | 0.004 | 0.4536 | No | ||
| 67 | RAB22A | 2888 5343 | 6581 | 0.004 | 0.4520 | No | ||
| 68 | FOXA1 | 21060 | 6822 | 0.004 | 0.4391 | No | ||
| 69 | GDF11 | 9011 | 6926 | 0.003 | 0.4336 | No | ||
| 70 | RAI2 | 10762 | 7170 | 0.003 | 0.4205 | No | ||
| 71 | DSC3 | 23487 | 7337 | 0.002 | 0.4115 | No | ||
| 72 | AMACR | 22506 | 7552 | 0.002 | 0.4000 | No | ||
| 73 | DAPK1 | 7632 | 7754 | 0.001 | 0.3891 | No | ||
| 74 | PTGFRN | 15224 | 7823 | 0.001 | 0.3854 | No | ||
| 75 | TIMP2 | 10178 | 8295 | -0.000 | 0.3600 | No | ||
| 76 | GUCY2C | 1140 16954 | 8573 | -0.001 | 0.3450 | No | ||
| 77 | SGCZ | 6354 | 8797 | -0.001 | 0.3329 | No | ||
| 78 | JAG1 | 14415 | 9053 | -0.002 | 0.3192 | No | ||
| 79 | ACTR3 | 13166 | 9295 | -0.003 | 0.3062 | No | ||
| 80 | NR3C2 | 18562 13798 | 9488 | -0.003 | 0.2958 | No | ||
| 81 | DDX42 | 20625 | 9912 | -0.004 | 0.2730 | No | ||
| 82 | WDR26 | 13733 | 9950 | -0.004 | 0.2711 | No | ||
| 83 | SET | 7070 | 10156 | -0.005 | 0.2601 | No | ||
| 84 | ANK2 | 4275 | 10185 | -0.005 | 0.2587 | No | ||
| 85 | ZIC1 | 19040 | 10535 | -0.006 | 0.2399 | No | ||
| 86 | ATP11A | 18683 6934 | 10735 | -0.006 | 0.2292 | No | ||
| 87 | DKK2 | 15418 | 10816 | -0.007 | 0.2250 | No | ||
| 88 | PABPN1 | 12018 | 10865 | -0.007 | 0.2225 | No | ||
| 89 | TAF4 | 14319 | 10882 | -0.007 | 0.2218 | No | ||
| 90 | TMEPAI | 51 | 11186 | -0.008 | 0.2056 | No | ||
| 91 | LRRN3 | 21077 | 11371 | -0.008 | 0.1958 | No | ||
| 92 | MYOG | 14129 | 11376 | -0.008 | 0.1957 | No | ||
| 93 | SATB2 | 5604 | 11417 | -0.009 | 0.1937 | No | ||
| 94 | CNN1 | 8759 | 11905 | -0.011 | 0.1676 | No | ||
| 95 | AFF2 | 8977 | 12063 | -0.011 | 0.1593 | No | ||
| 96 | PRKAB2 | 15488 | 12075 | -0.012 | 0.1589 | No | ||
| 97 | PLEKHM1 | 20196 1449 | 12287 | -0.013 | 0.1477 | No | ||
| 98 | PAX3 | 13910 | 12302 | -0.013 | 0.1472 | No | ||
| 99 | POU4F1 | 21727 | 12305 | -0.013 | 0.1474 | No | ||
| 100 | KCNJ2 | 20612 | 12394 | -0.013 | 0.1429 | No | ||
| 101 | ADRA2B | 8562 4356 14863 | 12528 | -0.014 | 0.1359 | No | ||
| 102 | PNN | 9596 | 12658 | -0.015 | 0.1292 | No | ||
| 103 | CREB3L2 | 17182 | 13011 | -0.018 | 0.1105 | No | ||
| 104 | PLCB1 | 14832 2821 | 13146 | -0.019 | 0.1036 | No | ||
| 105 | EIF3S1 | 905 8114 | 13180 | -0.019 | 0.1022 | No | ||
| 106 | BIRC4 | 2638 8609 | 13479 | -0.022 | 0.0865 | No | ||
| 107 | AEBP2 | 4359 | 13579 | -0.024 | 0.0816 | No | ||
| 108 | PHF6 | 2648 24340 | 13877 | -0.028 | 0.0661 | No | ||
| 109 | CATSPER2 | 10014 | 13893 | -0.028 | 0.0658 | No | ||
| 110 | DSCAML1 | 4338 13224 | 14064 | -0.032 | 0.0572 | No | ||
| 111 | PFN2 | 15337 1818 | 14090 | -0.032 | 0.0565 | No | ||
| 112 | PPM1F | 22841 1716 | 14208 | -0.035 | 0.0509 | No | ||
| 113 | RHOT1 | 12227 | 14241 | -0.035 | 0.0498 | No | ||
| 114 | STAG2 | 5521 | 14273 | -0.036 | 0.0488 | No | ||
| 115 | SP8 | 21122 | 14452 | -0.041 | 0.0400 | No | ||
| 116 | HMGA2 | 4857 9098 3341 3444 | 14519 | -0.043 | 0.0373 | No | ||
| 117 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 14527 | -0.043 | 0.0377 | No | ||
| 118 | YTHDF3 | 15630 10469 6019 | 14618 | -0.046 | 0.0337 | No | ||
| 119 | CPEB2 | 6077 | 14633 | -0.046 | 0.0339 | No | ||
| 120 | HNRPR | 13196 7909 2500 | 14697 | -0.048 | 0.0314 | No | ||
| 121 | DYRK1A | 4649 | 15249 | -0.076 | 0.0030 | No | ||
| 122 | CEPT1 | 13624 | 15283 | -0.078 | 0.0028 | No | ||
| 123 | KPNA4 | 15321 | 15473 | -0.092 | -0.0057 | No | ||
| 124 | TOMM70A | 6608 | 15480 | -0.093 | -0.0042 | No | ||
| 125 | EIF4G3 | 10517 | 15792 | -0.127 | -0.0186 | No | ||
| 126 | EGR3 | 4656 | 15854 | -0.136 | -0.0192 | No | ||
| 127 | HMGA1 | 23323 | 15876 | -0.140 | -0.0176 | No | ||
| 128 | CTDP1 | 23399 1969 | 16058 | -0.167 | -0.0242 | No | ||
| 129 | SEH1L | 13001 | 16077 | -0.171 | -0.0218 | No | ||
| 130 | EIF2C2 | 6247 | 16175 | -0.190 | -0.0234 | No | ||
| 131 | NRF1 | 17499 | 16351 | -0.228 | -0.0284 | No | ||
| 132 | DHX15 | 8842 | 16394 | -0.240 | -0.0260 | No | ||
| 133 | CORO1C | 16425 | 16397 | -0.242 | -0.0214 | No | ||
| 134 | PLEKHA3 | 14978 | 16441 | -0.252 | -0.0188 | No | ||
| 135 | KPNA3 | 21797 | 16540 | -0.279 | -0.0187 | No | ||
| 136 | RBBP7 | 10887 6366 10886 | 16809 | -0.348 | -0.0264 | No | ||
| 137 | PPP1R12A | 3354 19886 | 16837 | -0.357 | -0.0209 | No | ||
| 138 | RAB6A | 18173 3905 | 16923 | -0.384 | -0.0180 | No | ||
| 139 | NEDD8 | 21822 | 17156 | -0.462 | -0.0216 | No | ||
| 140 | NCKIPSD | 19306 | 17212 | -0.483 | -0.0152 | No | ||
| 141 | FUSIP1 | 4715 16036 | 17254 | -0.507 | -0.0075 | No | ||
| 142 | TARDBP | 10520 6066 | 17317 | -0.538 | -0.0004 | No | ||
| 143 | SLC25A26 | 12552 | 17677 | -0.730 | -0.0056 | No | ||
| 144 | UBAP2 | 15912 | 17817 | -0.826 | 0.0029 | No | ||
| 145 | ETF1 | 23467 | 17951 | -0.929 | 0.0138 | No | ||
| 146 | IVNS1ABP | 4384 4050 | 18124 | -1.135 | 0.0266 | No |