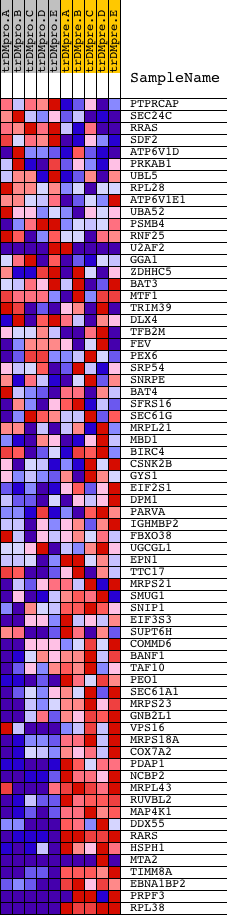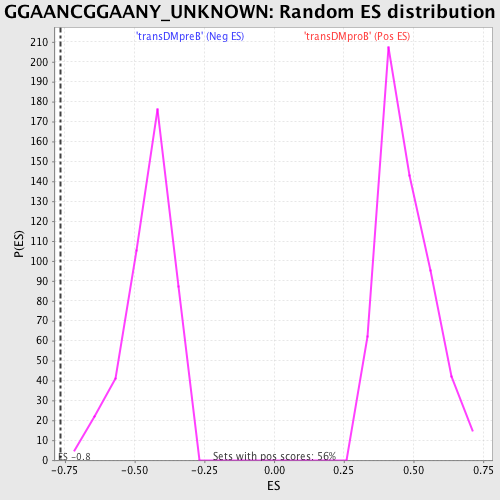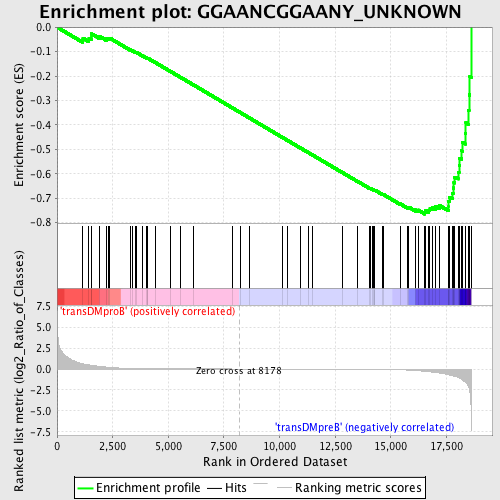
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | GGAANCGGAANY_UNKNOWN |
| Enrichment Score (ES) | -0.7650917 |
| Normalized Enrichment Score (NES) | -1.692929 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.013144604 |
| FWER p-Value | 0.031 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTPRCAP | 23767 | 1161 | 0.613 | -0.0452 | No | ||
| 2 | SEC24C | 22086 | 1431 | 0.490 | -0.0459 | No | ||
| 3 | RRAS | 18256 | 1543 | 0.442 | -0.0393 | No | ||
| 4 | SDF2 | 20762 | 1546 | 0.441 | -0.0270 | No | ||
| 5 | ATP6V1D | 7872 | 1901 | 0.314 | -0.0372 | No | ||
| 6 | PRKAB1 | 16408 | 2215 | 0.225 | -0.0477 | No | ||
| 7 | UBL5 | 12299 | 2296 | 0.207 | -0.0461 | No | ||
| 8 | RPL28 | 5392 | 2376 | 0.185 | -0.0451 | No | ||
| 9 | ATP6V1E1 | 4423 | 3292 | 0.063 | -0.0927 | No | ||
| 10 | UBA52 | 10239 | 3391 | 0.056 | -0.0964 | No | ||
| 11 | PSMB4 | 15252 | 3525 | 0.048 | -0.1022 | No | ||
| 12 | RNF25 | 13922 3929 | 3585 | 0.045 | -0.1041 | No | ||
| 13 | U2AF2 | 5813 5812 5814 | 3857 | 0.034 | -0.1177 | No | ||
| 14 | GGA1 | 22432 4197 | 3999 | 0.029 | -0.1245 | No | ||
| 15 | ZDHHC5 | 14547 | 4058 | 0.027 | -0.1269 | No | ||
| 16 | BAT3 | 5896 | 4074 | 0.027 | -0.1269 | No | ||
| 17 | MTF1 | 5128 9421 | 4407 | 0.019 | -0.1443 | No | ||
| 18 | TRIM39 | 22994 | 5082 | 0.011 | -0.1803 | No | ||
| 19 | DLX4 | 20282 1225 | 5566 | 0.008 | -0.2061 | No | ||
| 20 | TFB2M | 4024 9092 4083 | 6117 | 0.006 | -0.2356 | No | ||
| 21 | FEV | 11153 6434 | 7866 | 0.001 | -0.3298 | No | ||
| 22 | PEX6 | 5900 10331 | 8241 | -0.000 | -0.3500 | No | ||
| 23 | SRP54 | 6282 | 8649 | -0.001 | -0.3719 | No | ||
| 24 | SNRPE | 9843 | 10124 | -0.005 | -0.4512 | No | ||
| 25 | BAT4 | 8172 8173 | 10376 | -0.005 | -0.4646 | No | ||
| 26 | SFRS16 | 17943 3840 | 10928 | -0.007 | -0.4941 | No | ||
| 27 | SEC61G | 9795 | 11279 | -0.008 | -0.5127 | No | ||
| 28 | MRPL21 | 23755 | 11463 | -0.009 | -0.5224 | No | ||
| 29 | MBD1 | 1959 23518 1998 2020 | 12835 | -0.016 | -0.5958 | No | ||
| 30 | BIRC4 | 2638 8609 | 13479 | -0.022 | -0.6299 | No | ||
| 31 | CSNK2B | 23008 | 14052 | -0.031 | -0.6598 | No | ||
| 32 | GYS1 | 4818 | 14075 | -0.032 | -0.6601 | No | ||
| 33 | EIF2S1 | 4658 | 14189 | -0.034 | -0.6652 | No | ||
| 34 | DPM1 | 4636 2882 8860 | 14237 | -0.035 | -0.6667 | No | ||
| 35 | PARVA | 18122 | 14258 | -0.036 | -0.6668 | No | ||
| 36 | IGHMBP2 | 23945 | 14285 | -0.036 | -0.6672 | No | ||
| 37 | FBXO38 | 23421 | 14616 | -0.045 | -0.6837 | No | ||
| 38 | UGCGL1 | 6721 | 14655 | -0.047 | -0.6844 | No | ||
| 39 | EPN1 | 18401 | 15413 | -0.087 | -0.7228 | No | ||
| 40 | TTC17 | 13219 | 15751 | -0.122 | -0.7375 | No | ||
| 41 | MRPS21 | 12323 | 15804 | -0.129 | -0.7366 | No | ||
| 42 | SMUG1 | 22109 | 16111 | -0.179 | -0.7481 | No | ||
| 43 | SNIP1 | 16085 | 16224 | -0.201 | -0.7484 | No | ||
| 44 | EIF3S3 | 12652 | 16534 | -0.277 | -0.7572 | Yes | ||
| 45 | SUPT6H | 9940 | 16559 | -0.283 | -0.7505 | Yes | ||
| 46 | COMMD6 | 476 | 16709 | -0.322 | -0.7494 | Yes | ||
| 47 | BANF1 | 6216 10706 3760 | 16759 | -0.333 | -0.7426 | Yes | ||
| 48 | TAF10 | 10785 | 16868 | -0.367 | -0.7381 | Yes | ||
| 49 | PEO1 | 5932 | 17017 | -0.416 | -0.7343 | Yes | ||
| 50 | SEC61A1 | 60 | 17194 | -0.477 | -0.7303 | Yes | ||
| 51 | MRPS23 | 12260 | 17595 | -0.686 | -0.7324 | Yes | ||
| 52 | GNB2L1 | 20911 | 17600 | -0.690 | -0.7131 | Yes | ||
| 53 | VPS16 | 2927 14845 2825 | 17644 | -0.709 | -0.6953 | Yes | ||
| 54 | MRPS18A | 23222 | 17769 | -0.794 | -0.6796 | Yes | ||
| 55 | COX7A2 | 19053 | 17802 | -0.817 | -0.6582 | Yes | ||
| 56 | PDAP1 | 16294 | 17834 | -0.842 | -0.6360 | Yes | ||
| 57 | NCBP2 | 12643 | 17873 | -0.871 | -0.6134 | Yes | ||
| 58 | MRPL43 | 3703 23661 | 18025 | -1.004 | -0.5931 | Yes | ||
| 59 | RUVBL2 | 9766 | 18069 | -1.062 | -0.5654 | Yes | ||
| 60 | MAP4K1 | 18313 | 18098 | -1.095 | -0.5359 | Yes | ||
| 61 | DDX55 | 7490 | 18181 | -1.215 | -0.5059 | Yes | ||
| 62 | RARS | 20496 | 18210 | -1.272 | -0.4714 | Yes | ||
| 63 | HSPH1 | 16282 | 18341 | -1.529 | -0.4352 | Yes | ||
| 64 | MTA2 | 23934 18250 | 18374 | -1.602 | -0.3915 | Yes | ||
| 65 | TIMM8A | 24062 | 18478 | -2.024 | -0.3398 | Yes | ||
| 66 | EBNA1BP2 | 8112 7599 12770 | 18517 | -2.300 | -0.2768 | Yes | ||
| 67 | PRPF3 | 12899 12898 1877 7703 | 18551 | -2.691 | -0.2024 | Yes | ||
| 68 | RPL38 | 12562 20606 7475 | 18616 | -7.271 | -0.0000 | Yes |