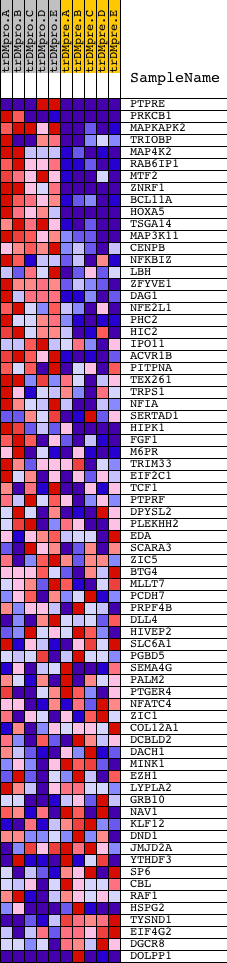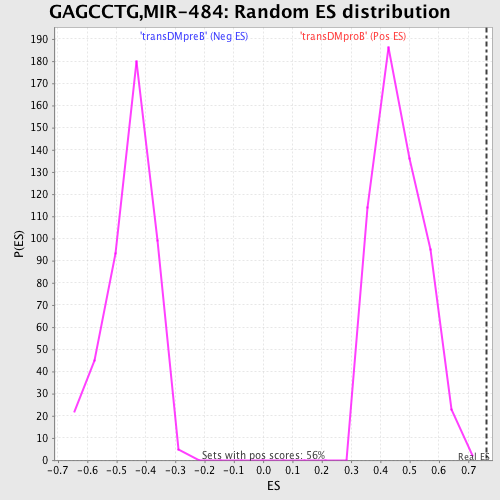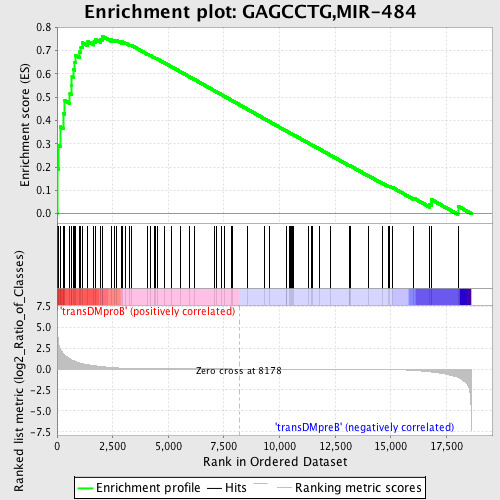
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | GAGCCTG,MIR-484 |
| Enrichment Score (ES) | 0.76209384 |
| Normalized Enrichment Score (NES) | 1.6413313 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029675988 |
| FWER p-Value | 0.114 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTPRE | 1662 18039 | 4 | 5.427 | 0.1939 | Yes | ||
| 2 | PRKCB1 | 1693 9574 | 81 | 2.858 | 0.2921 | Yes | ||
| 3 | MAPKAPK2 | 13838 | 146 | 2.333 | 0.3721 | Yes | ||
| 4 | TRIOBP | 2280 670 8476 | 272 | 1.836 | 0.4311 | Yes | ||
| 5 | MAP4K2 | 6457 | 348 | 1.618 | 0.4849 | Yes | ||
| 6 | RAB6IP1 | 17676 | 577 | 1.204 | 0.5157 | Yes | ||
| 7 | MTF2 | 5129 | 664 | 1.068 | 0.5493 | Yes | ||
| 8 | ZNRF1 | 9306 | 668 | 1.064 | 0.5872 | Yes | ||
| 9 | BCL11A | 4691 | 748 | 0.967 | 0.6175 | Yes | ||
| 10 | HOXA5 | 17149 | 799 | 0.916 | 0.6476 | Yes | ||
| 11 | TSGA14 | 17194 | 815 | 0.904 | 0.6791 | Yes | ||
| 12 | MAP3K11 | 11163 | 1013 | 0.714 | 0.6940 | Yes | ||
| 13 | CENPB | 14424 | 1070 | 0.667 | 0.7149 | Yes | ||
| 14 | NFKBIZ | 22575 | 1140 | 0.623 | 0.7335 | Yes | ||
| 15 | LBH | 23160 | 1383 | 0.510 | 0.7386 | Yes | ||
| 16 | ZFYVE1 | 10138 | 1657 | 0.395 | 0.7380 | Yes | ||
| 17 | DAG1 | 18996 8837 | 1712 | 0.374 | 0.7485 | Yes | ||
| 18 | NFE2L1 | 9457 | 1952 | 0.301 | 0.7464 | Yes | ||
| 19 | PHC2 | 16075 | 2026 | 0.278 | 0.7524 | Yes | ||
| 20 | HIC2 | 7185 | 2030 | 0.277 | 0.7621 | Yes | ||
| 21 | IPO11 | 21353 | 2445 | 0.173 | 0.7460 | No | ||
| 22 | ACVR1B | 4335 22353 | 2599 | 0.145 | 0.7429 | No | ||
| 23 | PITPNA | 20778 | 2677 | 0.133 | 0.7435 | No | ||
| 24 | TEX261 | 17094 5726 | 2891 | 0.101 | 0.7356 | No | ||
| 25 | TRPS1 | 8195 | 2925 | 0.098 | 0.7373 | No | ||
| 26 | NFIA | 16172 5170 | 3093 | 0.082 | 0.7313 | No | ||
| 27 | SERTAD1 | 18326 | 3274 | 0.065 | 0.7239 | No | ||
| 28 | HIPK1 | 4851 | 3337 | 0.061 | 0.7227 | No | ||
| 29 | FGF1 | 1994 23447 | 4054 | 0.027 | 0.6851 | No | ||
| 30 | M6PR | 1053 5044 9328 | 4193 | 0.024 | 0.6785 | No | ||
| 31 | TRIM33 | 8236 13579 15472 8237 | 4377 | 0.019 | 0.6693 | No | ||
| 32 | EIF2C1 | 10672 | 4422 | 0.018 | 0.6676 | No | ||
| 33 | TCF1 | 16416 | 4523 | 0.017 | 0.6628 | No | ||
| 34 | PTPRF | 5331 2545 | 4823 | 0.013 | 0.6472 | No | ||
| 35 | DPYSL2 | 3122 4561 8787 | 5140 | 0.011 | 0.6305 | No | ||
| 36 | PLEKHH2 | 23148 | 5557 | 0.008 | 0.6084 | No | ||
| 37 | EDA | 24284 | 5937 | 0.007 | 0.5882 | No | ||
| 38 | SCARA3 | 21780 | 6192 | 0.006 | 0.5747 | No | ||
| 39 | ZIC5 | 12268 | 7084 | 0.003 | 0.5267 | No | ||
| 40 | BTG4 | 19456 | 7156 | 0.003 | 0.5230 | No | ||
| 41 | MLLT7 | 24278 | 7398 | 0.002 | 0.5101 | No | ||
| 42 | PCDH7 | 7029 3460 | 7522 | 0.002 | 0.5035 | No | ||
| 43 | PRPF4B | 3208 5295 | 7854 | 0.001 | 0.4857 | No | ||
| 44 | DLL4 | 7040 | 7865 | 0.001 | 0.4852 | No | ||
| 45 | HIVEP2 | 9091 | 8577 | -0.001 | 0.4469 | No | ||
| 46 | SLC6A1 | 10557 | 9303 | -0.003 | 0.4079 | No | ||
| 47 | PGBD5 | 9960 | 9547 | -0.003 | 0.3949 | No | ||
| 48 | SEMA4G | 11182 | 10293 | -0.005 | 0.3549 | No | ||
| 49 | PALM2 | 10817 2368 | 10300 | -0.005 | 0.3547 | No | ||
| 50 | PTGER4 | 9649 | 10435 | -0.005 | 0.3477 | No | ||
| 51 | NFATC4 | 22002 | 10475 | -0.006 | 0.3458 | No | ||
| 52 | ZIC1 | 19040 | 10535 | -0.006 | 0.3428 | No | ||
| 53 | COL12A1 | 3143 19054 | 10588 | -0.006 | 0.3402 | No | ||
| 54 | DCBLD2 | 7855 | 10636 | -0.006 | 0.3379 | No | ||
| 55 | DACH1 | 4595 3371 | 11316 | -0.008 | 0.3016 | No | ||
| 56 | MINK1 | 20802 | 11444 | -0.009 | 0.2950 | No | ||
| 57 | EZH1 | 20217 | 11467 | -0.009 | 0.2942 | No | ||
| 58 | LYPLA2 | 15708 | 11777 | -0.010 | 0.2779 | No | ||
| 59 | GRB10 | 4799 | 12283 | -0.013 | 0.2511 | No | ||
| 60 | NAV1 | 3978 5681 | 13148 | -0.019 | 0.2052 | No | ||
| 61 | KLF12 | 4960 2637 21732 9228 | 13156 | -0.019 | 0.2055 | No | ||
| 62 | DND1 | 10017 | 13178 | -0.019 | 0.2050 | No | ||
| 63 | JMJD2A | 10505 6058 | 14001 | -0.031 | 0.1618 | No | ||
| 64 | YTHDF3 | 15630 10469 6019 | 14618 | -0.046 | 0.1302 | No | ||
| 65 | SP6 | 8178 13678 13679 | 14878 | -0.055 | 0.1182 | No | ||
| 66 | CBL | 19154 | 14950 | -0.058 | 0.1165 | No | ||
| 67 | RAF1 | 17035 | 15064 | -0.065 | 0.1127 | No | ||
| 68 | HSPG2 | 4884 16023 | 16031 | -0.164 | 0.0664 | No | ||
| 69 | TYSND1 | 20007 | 16744 | -0.331 | 0.0399 | No | ||
| 70 | EIF4G2 | 1908 8892 | 16813 | -0.350 | 0.0487 | No | ||
| 71 | DGCR8 | 22644 | 16820 | -0.351 | 0.0609 | No | ||
| 72 | DOLPP1 | 2682 15051 | 18020 | -1.002 | 0.0321 | No |