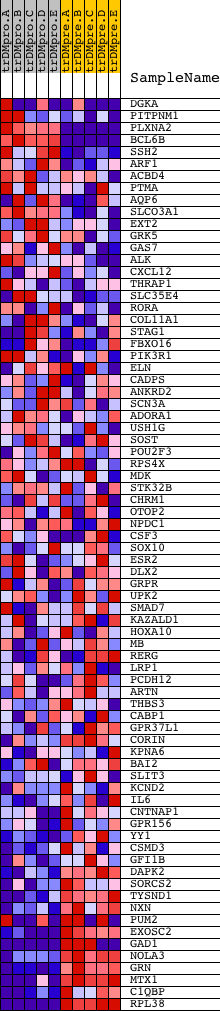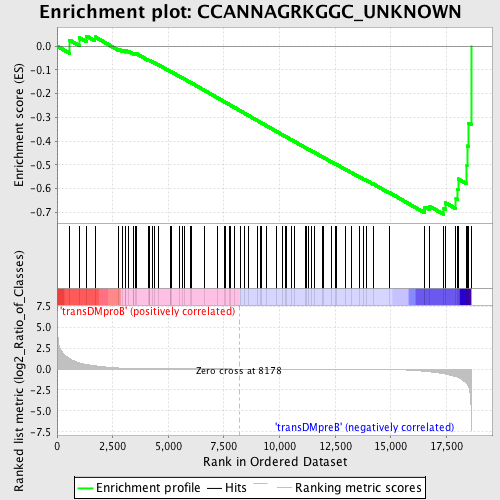
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | CCANNAGRKGGC_UNKNOWN |
| Enrichment Score (ES) | -0.70842195 |
| Normalized Enrichment Score (NES) | -1.5903487 |
| Nominal p-value | 0.0023148148 |
| FDR q-value | 0.065413855 |
| FWER p-Value | 0.267 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DGKA | 3359 19589 | 563 | 1.224 | 0.0252 | No | ||
| 2 | PITPNM1 | 23770 | 990 | 0.743 | 0.0359 | No | ||
| 3 | PLXNA2 | 5269 | 1326 | 0.534 | 0.0421 | No | ||
| 4 | BCL6B | 20373 | 1703 | 0.379 | 0.0389 | No | ||
| 5 | SSH2 | 6202 | 2764 | 0.118 | -0.0129 | No | ||
| 6 | ARF1 | 64 | 2956 | 0.095 | -0.0189 | No | ||
| 7 | ACBD4 | 20637 | 3069 | 0.084 | -0.0211 | No | ||
| 8 | PTMA | 9657 | 3085 | 0.082 | -0.0182 | No | ||
| 9 | AQP6 | 22364 8621 | 3221 | 0.069 | -0.0223 | No | ||
| 10 | SLCO3A1 | 17796 4237 8430 | 3429 | 0.054 | -0.0310 | No | ||
| 11 | EXT2 | 14516 | 3440 | 0.054 | -0.0291 | No | ||
| 12 | GRK5 | 9036 23814 | 3534 | 0.048 | -0.0320 | No | ||
| 13 | GAS7 | 20836 1299 | 3552 | 0.047 | -0.0308 | No | ||
| 14 | ALK | 8574 | 4094 | 0.026 | -0.0588 | No | ||
| 15 | CXCL12 | 9792 1182 1031 | 4155 | 0.024 | -0.0609 | No | ||
| 16 | THRAP1 | 20309 | 4157 | 0.024 | -0.0599 | No | ||
| 17 | SLC35E4 | 20549 | 4305 | 0.021 | -0.0669 | No | ||
| 18 | RORA | 3019 5388 | 4394 | 0.019 | -0.0708 | No | ||
| 19 | COL11A1 | 15442 1937 4540 8762 | 4544 | 0.017 | -0.0780 | No | ||
| 20 | STAG1 | 5520 9905 2987 | 5091 | 0.011 | -0.1070 | No | ||
| 21 | FBXO16 | 11929 | 5107 | 0.011 | -0.1073 | No | ||
| 22 | PIK3R1 | 3170 | 5127 | 0.011 | -0.1078 | No | ||
| 23 | ELN | 8896 | 5481 | 0.009 | -0.1265 | No | ||
| 24 | CADPS | 11298 | 5617 | 0.008 | -0.1334 | No | ||
| 25 | ANKRD2 | 23853 | 5715 | 0.007 | -0.1383 | No | ||
| 26 | SCN3A | 14573 | 5986 | 0.006 | -0.1526 | No | ||
| 27 | ADORA1 | 13831 | 6024 | 0.006 | -0.1543 | No | ||
| 28 | USH1G | 20153 | 6616 | 0.004 | -0.1860 | No | ||
| 29 | SOST | 20210 | 7209 | 0.003 | -0.2178 | No | ||
| 30 | POU2F3 | 9602 | 7524 | 0.002 | -0.2346 | No | ||
| 31 | RPS4X | 9756 | 7586 | 0.002 | -0.2379 | No | ||
| 32 | MDK | 14523 | 7769 | 0.001 | -0.2476 | No | ||
| 33 | STK32B | 12255 | 7814 | 0.001 | -0.2500 | No | ||
| 34 | CHRM1 | 23943 | 7972 | 0.001 | -0.2584 | No | ||
| 35 | OTOP2 | 20600 | 8220 | -0.000 | -0.2717 | No | ||
| 36 | NPDC1 | 9479 | 8406 | -0.001 | -0.2817 | No | ||
| 37 | CSF3 | 1394 20671 | 8594 | -0.001 | -0.2917 | No | ||
| 38 | SOX10 | 22211 | 9004 | -0.002 | -0.3137 | No | ||
| 39 | ESR2 | 21038 | 9139 | -0.002 | -0.3208 | No | ||
| 40 | DLX2 | 14561 353 | 9164 | -0.002 | -0.3220 | No | ||
| 41 | GRPR | 24015 | 9429 | -0.003 | -0.3361 | No | ||
| 42 | UPK2 | 19146 | 9880 | -0.004 | -0.3602 | No | ||
| 43 | SMAD7 | 23513 | 10120 | -0.005 | -0.3729 | No | ||
| 44 | KAZALD1 | 4206 | 10275 | -0.005 | -0.3810 | No | ||
| 45 | HOXA10 | 17145 989 | 10319 | -0.005 | -0.3831 | No | ||
| 46 | MB | 22232 | 10518 | -0.006 | -0.3935 | No | ||
| 47 | RERG | 16949 | 10680 | -0.006 | -0.4019 | No | ||
| 48 | LRP1 | 9284 | 11148 | -0.008 | -0.4267 | No | ||
| 49 | PCDH12 | 7005 | 11221 | -0.008 | -0.4303 | No | ||
| 50 | ARTN | 15783 2430 2532 | 11301 | -0.008 | -0.4342 | No | ||
| 51 | THBS3 | 15542 1796 | 11449 | -0.009 | -0.4417 | No | ||
| 52 | CABP1 | 16412 | 11571 | -0.009 | -0.4478 | No | ||
| 53 | GPR37L1 | 13825 | 11939 | -0.011 | -0.4671 | No | ||
| 54 | CORIN | 16517 | 11966 | -0.011 | -0.4680 | No | ||
| 55 | KPNA6 | 4972 | 12345 | -0.013 | -0.4878 | No | ||
| 56 | BAI2 | 2541 2379 16067 | 12521 | -0.014 | -0.4966 | No | ||
| 57 | SLIT3 | 20925 | 12553 | -0.014 | -0.4976 | No | ||
| 58 | KCND2 | 4941 17515 | 12955 | -0.017 | -0.5185 | No | ||
| 59 | IL6 | 16895 | 13211 | -0.019 | -0.5313 | No | ||
| 60 | CNTNAP1 | 11987 | 13584 | -0.024 | -0.5503 | No | ||
| 61 | GPR156 | 10753 22763 | 13786 | -0.027 | -0.5600 | No | ||
| 62 | YY1 | 10371 | 13908 | -0.029 | -0.5652 | No | ||
| 63 | CSMD3 | 10735 7938 | 14203 | -0.034 | -0.5795 | No | ||
| 64 | GFI1B | 14637 | 14921 | -0.057 | -0.6156 | No | ||
| 65 | DAPK2 | 19395 | 16519 | -0.271 | -0.6894 | Yes | ||
| 66 | SORCS2 | 16553 | 16528 | -0.275 | -0.6774 | Yes | ||
| 67 | TYSND1 | 20007 | 16744 | -0.331 | -0.6740 | Yes | ||
| 68 | NXN | 5204 | 17384 | -0.570 | -0.6825 | Yes | ||
| 69 | PUM2 | 2071 8161 | 17457 | -0.602 | -0.6591 | Yes | ||
| 70 | EXOSC2 | 15044 | 17927 | -0.912 | -0.6430 | Yes | ||
| 71 | GAD1 | 14993 2690 | 17985 | -0.960 | -0.6026 | Yes | ||
| 72 | NOLA3 | 14914 | 18031 | -1.015 | -0.5590 | Yes | ||
| 73 | GRN | 20640 | 18421 | -1.752 | -0.5005 | Yes | ||
| 74 | MTX1 | 15282 1918 | 18427 | -1.779 | -0.4201 | Yes | ||
| 75 | C1QBP | 20364 | 18500 | -2.215 | -0.3236 | Yes | ||
| 76 | RPL38 | 12562 20606 7475 | 18616 | -7.271 | 0.0000 | Yes |