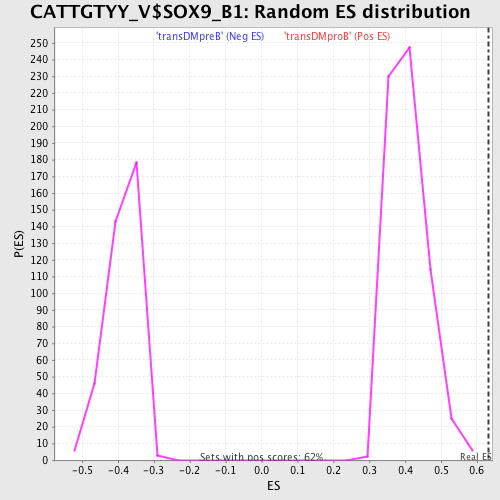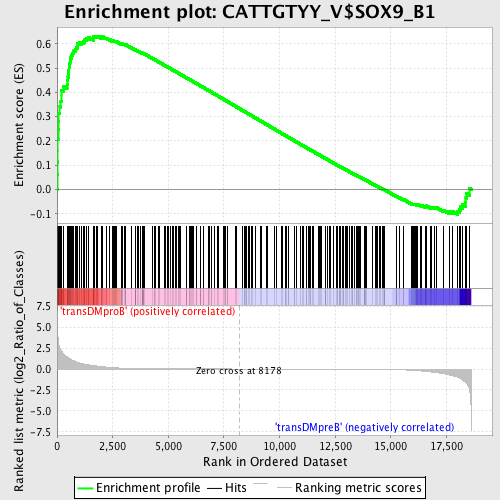
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | CATTGTYY_V$SOX9_B1 |
| Enrichment Score (ES) | 0.63177395 |
| Normalized Enrichment Score (NES) | 1.5524963 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07013591 |
| FWER p-Value | 0.506 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 8214 2501 13554 | 6 | 5.419 | 0.0619 | Yes | ||
| 2 | RANBP1 | 9692 5357 | 14 | 4.554 | 0.1138 | Yes | ||
| 3 | DAB2IP | 12808 2692 | 26 | 4.153 | 0.1609 | Yes | ||
| 4 | GFRA1 | 9015 | 28 | 4.130 | 0.2082 | Yes | ||
| 5 | CDKN1A | 4511 8729 | 49 | 3.556 | 0.2479 | Yes | ||
| 6 | MARCKS | 9331 | 69 | 3.057 | 0.2820 | Yes | ||
| 7 | TPM4 | 11589 11588 6819 | 78 | 2.914 | 0.3150 | Yes | ||
| 8 | TIAM1 | 22547 | 127 | 2.451 | 0.3405 | Yes | ||
| 9 | CNN3 | 12983 | 168 | 2.250 | 0.3642 | Yes | ||
| 10 | HMGN3 | 19050 3091 | 208 | 2.074 | 0.3859 | Yes | ||
| 11 | MAML1 | 20477 | 209 | 2.068 | 0.4096 | Yes | ||
| 12 | TCF4 | 5642 | 292 | 1.758 | 0.4253 | Yes | ||
| 13 | MEIS1 | 20524 | 446 | 1.464 | 0.4338 | Yes | ||
| 14 | RRM2 | 5401 5400 | 461 | 1.431 | 0.4495 | Yes | ||
| 15 | MYLK | 22778 4213 | 488 | 1.366 | 0.4637 | Yes | ||
| 16 | DLX1 | 14988 | 502 | 1.333 | 0.4783 | Yes | ||
| 17 | HHEX | 23872 | 526 | 1.273 | 0.4917 | Yes | ||
| 18 | STAT3 | 5525 9906 | 534 | 1.263 | 0.5058 | Yes | ||
| 19 | RASSF2 | 2802 14422 | 562 | 1.225 | 0.5184 | Yes | ||
| 20 | HSD11B1 | 9125 4019 | 581 | 1.194 | 0.5311 | Yes | ||
| 21 | DDAH2 | 23263 | 616 | 1.122 | 0.5422 | Yes | ||
| 22 | CLIC4 | 11386 11342 6620 11387 | 636 | 1.102 | 0.5538 | Yes | ||
| 23 | P2RX3 | 10446 | 703 | 1.017 | 0.5619 | Yes | ||
| 24 | BCL11A | 4691 | 748 | 0.967 | 0.5706 | Yes | ||
| 25 | S100A1 | 15271 | 823 | 0.898 | 0.5768 | Yes | ||
| 26 | SLC4A1 | 20204 | 851 | 0.876 | 0.5854 | Yes | ||
| 27 | PRKCH | 21246 | 909 | 0.818 | 0.5917 | Yes | ||
| 28 | ELOVL6 | 5009 | 918 | 0.805 | 0.6005 | Yes | ||
| 29 | H3F3B | 4839 | 993 | 0.739 | 0.6050 | Yes | ||
| 30 | ABCF2 | 11338 6580 6579 | 1103 | 0.651 | 0.6065 | Yes | ||
| 31 | RBMS1 | 14580 | 1190 | 0.598 | 0.6087 | Yes | ||
| 32 | TRAF4 | 10217 5796 1400 | 1244 | 0.572 | 0.6124 | Yes | ||
| 33 | RAB1A | 20942 | 1249 | 0.569 | 0.6187 | Yes | ||
| 34 | ITCH | 4928 9185 | 1329 | 0.533 | 0.6205 | Yes | ||
| 35 | NT5C2 | 3768 8052 | 1402 | 0.501 | 0.6223 | Yes | ||
| 36 | NDST2 | 21906 | 1422 | 0.493 | 0.6270 | Yes | ||
| 37 | VCL | 22083 | 1641 | 0.401 | 0.6197 | Yes | ||
| 38 | RANBP9 | 21480 | 1642 | 0.400 | 0.6243 | Yes | ||
| 39 | BMF | 5074 | 1645 | 0.400 | 0.6288 | Yes | ||
| 40 | MAP4K4 | 11241 | 1693 | 0.381 | 0.6306 | Yes | ||
| 41 | PHC1 | 17013 | 1787 | 0.351 | 0.6295 | Yes | ||
| 42 | MYO18A | 20766 | 1818 | 0.336 | 0.6318 | Yes | ||
| 43 | ADAM15 | 15275 | 1978 | 0.292 | 0.6265 | No | ||
| 44 | FBXL20 | 13010 | 2002 | 0.283 | 0.6285 | No | ||
| 45 | PICALM | 18191 | 2061 | 0.266 | 0.6284 | No | ||
| 46 | EPHB6 | 17473 | 2208 | 0.226 | 0.6230 | No | ||
| 47 | GPC2 | 16321 | 2368 | 0.187 | 0.6165 | No | ||
| 48 | PPP1R16B | 6014 | 2477 | 0.164 | 0.6125 | No | ||
| 49 | EIF4EBP2 | 4662 | 2523 | 0.157 | 0.6119 | No | ||
| 50 | CDK2 | 3438 3373 19592 3322 | 2570 | 0.150 | 0.6111 | No | ||
| 51 | HN1 | 9101 | 2607 | 0.144 | 0.6108 | No | ||
| 52 | MBP | 23502 | 2669 | 0.134 | 0.6090 | No | ||
| 53 | DMD | 24295 2647 | 2886 | 0.102 | 0.5984 | No | ||
| 54 | POLB | 9599 | 2898 | 0.101 | 0.5990 | No | ||
| 55 | TOP1 | 5790 5789 10210 | 2904 | 0.100 | 0.5998 | No | ||
| 56 | TBX6 | 1993 18075 | 2940 | 0.096 | 0.5990 | No | ||
| 57 | ELAVL1 | 4888 | 2945 | 0.096 | 0.5999 | No | ||
| 58 | SEPHS1 | 15125 | 3018 | 0.088 | 0.5970 | No | ||
| 59 | APRIN | 4145 | 3057 | 0.085 | 0.5959 | No | ||
| 60 | PTMA | 9657 | 3085 | 0.082 | 0.5954 | No | ||
| 61 | KIF1B | 15671 2476 4950 15673 | 3088 | 0.082 | 0.5962 | No | ||
| 62 | SLC39A9 | 11597 | 3328 | 0.061 | 0.5839 | No | ||
| 63 | SLC26A9 | 11560 980 4067 | 3510 | 0.049 | 0.5746 | No | ||
| 64 | CD68 | 4501 | 3523 | 0.048 | 0.5745 | No | ||
| 65 | GRK5 | 9036 23814 | 3534 | 0.048 | 0.5745 | No | ||
| 66 | CD96 | 22580 | 3536 | 0.048 | 0.5750 | No | ||
| 67 | TLE1 | 5765 10185 15859 | 3627 | 0.043 | 0.5706 | No | ||
| 68 | SOX5 | 9850 1044 5480 16937 | 3677 | 0.041 | 0.5684 | No | ||
| 69 | ANK3 | 8591 3445 3304 3381 | 3750 | 0.038 | 0.5649 | No | ||
| 70 | PBX1 | 5224 | 3815 | 0.036 | 0.5618 | No | ||
| 71 | SIRT6 | 19668 3447 | 3816 | 0.036 | 0.5622 | No | ||
| 72 | NOTCH3 | 23031 | 3821 | 0.035 | 0.5624 | No | ||
| 73 | RAPH1 | 19110 | 3845 | 0.034 | 0.5616 | No | ||
| 74 | GPR85 | 7227 1048 | 3865 | 0.034 | 0.5609 | No | ||
| 75 | OLFM1 | 2918 2971 | 3870 | 0.033 | 0.5611 | No | ||
| 76 | NUDT3 | 23056 | 3901 | 0.032 | 0.5598 | No | ||
| 77 | CD151 | 18010 4494 | 3929 | 0.031 | 0.5587 | No | ||
| 78 | MYCT1 | 7572 | 4282 | 0.021 | 0.5398 | No | ||
| 79 | LIN28 | 15723 | 4361 | 0.020 | 0.5358 | No | ||
| 80 | TNPO1 | 6232 3179 | 4381 | 0.019 | 0.5350 | No | ||
| 81 | VWF | 17277 | 4433 | 0.018 | 0.5324 | No | ||
| 82 | MAPK3 | 6458 11170 | 4552 | 0.016 | 0.5262 | No | ||
| 83 | CORO1A | 1066 17631 1074 | 4597 | 0.016 | 0.5239 | No | ||
| 84 | FBXO28 | 7508 12612 | 4813 | 0.013 | 0.5124 | No | ||
| 85 | GNG11 | 17532 | 4860 | 0.013 | 0.5100 | No | ||
| 86 | CNTF | 3741 8761 3680 3736 | 4979 | 0.012 | 0.5037 | No | ||
| 87 | CALD1 | 4273 8463 | 5005 | 0.012 | 0.5025 | No | ||
| 88 | SILV | 16130 | 5081 | 0.011 | 0.4986 | No | ||
| 89 | CFL2 | 2155 21069 | 5194 | 0.010 | 0.4926 | No | ||
| 90 | DCX | 24040 8841 4604 | 5228 | 0.010 | 0.4909 | No | ||
| 91 | APLN | 24164 | 5329 | 0.010 | 0.4856 | No | ||
| 92 | MITF | 17349 | 5365 | 0.009 | 0.4838 | No | ||
| 93 | BCL2L1 | 4440 2930 8652 | 5474 | 0.009 | 0.4780 | No | ||
| 94 | FGF9 | 8966 | 5506 | 0.009 | 0.4764 | No | ||
| 95 | MAOB | 24182 2556 | 5558 | 0.008 | 0.4737 | No | ||
| 96 | DOCK4 | 10705 | 5814 | 0.007 | 0.4599 | No | ||
| 97 | SMARCD1 | 2241 22360 2250 | 5823 | 0.007 | 0.4596 | No | ||
| 98 | HMGB1 | 9094 4855 | 5826 | 0.007 | 0.4595 | No | ||
| 99 | MEOX1 | 20211 | 5938 | 0.007 | 0.4536 | No | ||
| 100 | BMP5 | 19376 | 5941 | 0.007 | 0.4535 | No | ||
| 101 | GNAO1 | 4785 3829 | 6003 | 0.006 | 0.4503 | No | ||
| 102 | PNOC | 21783 | 6023 | 0.006 | 0.4493 | No | ||
| 103 | TRIM2 | 13485 8157 | 6074 | 0.006 | 0.4467 | No | ||
| 104 | MYL3 | 19291 | 6142 | 0.006 | 0.4431 | No | ||
| 105 | OSBP2 | 13194 | 6267 | 0.005 | 0.4364 | No | ||
| 106 | CHN1 | 4246 | 6272 | 0.005 | 0.4362 | No | ||
| 107 | LAMA1 | 23168 | 6455 | 0.005 | 0.4264 | No | ||
| 108 | BAMBI | 23629 | 6458 | 0.005 | 0.4263 | No | ||
| 109 | CLOCK | 8749 16507 4530 | 6460 | 0.005 | 0.4263 | No | ||
| 110 | CHL1 | 17347 1146 8741 4520 | 6564 | 0.004 | 0.4208 | No | ||
| 111 | CHST8 | 12754 | 6587 | 0.004 | 0.4196 | No | ||
| 112 | FOXA1 | 21060 | 6822 | 0.004 | 0.4069 | No | ||
| 113 | PNRC1 | 15925 | 6825 | 0.004 | 0.4068 | No | ||
| 114 | PTPN4 | 13856 | 6852 | 0.003 | 0.4055 | No | ||
| 115 | SYTL2 | 18190 1539 3730 | 6956 | 0.003 | 0.3999 | No | ||
| 116 | HOXA3 | 9108 1075 | 6958 | 0.003 | 0.3999 | No | ||
| 117 | TIA1 | 5752 | 7081 | 0.003 | 0.3933 | No | ||
| 118 | TREX1 | 10219 3111 | 7207 | 0.003 | 0.3865 | No | ||
| 119 | SRGAP1 | 19615 | 7247 | 0.002 | 0.3844 | No | ||
| 120 | VLDLR | 5858 3763 912 3682 | 7469 | 0.002 | 0.3724 | No | ||
| 121 | NKX2-2 | 5176 | 7544 | 0.002 | 0.3684 | No | ||
| 122 | SPG3A | 7878 13145 | 7551 | 0.002 | 0.3681 | No | ||
| 123 | HOXA2 | 17151 | 7568 | 0.002 | 0.3672 | No | ||
| 124 | PAFAH1B2 | 5221 9525 | 7653 | 0.001 | 0.3627 | No | ||
| 125 | AP1S2 | 8423 4228 | 7671 | 0.001 | 0.3617 | No | ||
| 126 | ZNF8 | 6339 | 8005 | 0.000 | 0.3436 | No | ||
| 127 | NGFR | 5174 | 8055 | 0.000 | 0.3410 | No | ||
| 128 | MTSS1 | 5585 22285 | 8332 | -0.000 | 0.3259 | No | ||
| 129 | SMARCA5 | 8215 | 8411 | -0.001 | 0.3217 | No | ||
| 130 | BASP1 | 7668 | 8425 | -0.001 | 0.3210 | No | ||
| 131 | NDST3 | 13501 | 8466 | -0.001 | 0.3188 | No | ||
| 132 | PRDM8 | 8084 | 8501 | -0.001 | 0.3170 | No | ||
| 133 | OLIG3 | 20084 | 8527 | -0.001 | 0.3156 | No | ||
| 134 | CSF3 | 1394 20671 | 8594 | -0.001 | 0.3120 | No | ||
| 135 | SIX1 | 9821 | 8647 | -0.001 | 0.3092 | No | ||
| 136 | CDX1 | 23429 | 8749 | -0.001 | 0.3037 | No | ||
| 137 | PER2 | 3940 13881 | 8769 | -0.001 | 0.3027 | No | ||
| 138 | MYH2 | 20838 | 8935 | -0.002 | 0.2938 | No | ||
| 139 | MOSPD2 | 24009 | 9146 | -0.002 | 0.2823 | No | ||
| 140 | NEUROD2 | 20262 | 9167 | -0.002 | 0.2813 | No | ||
| 141 | OLIG2 | 6953 | 9428 | -0.003 | 0.2672 | No | ||
| 142 | LHX6 | 9275 2914 | 9464 | -0.003 | 0.2653 | No | ||
| 143 | HOXD3 | 9115 | 9774 | -0.004 | 0.2485 | No | ||
| 144 | PRKCQ | 2873 2831 | 9877 | -0.004 | 0.2430 | No | ||
| 145 | ARHGEF19 | 10035 | 10104 | -0.005 | 0.2307 | No | ||
| 146 | KCNN2 | 8933 1949 | 10148 | -0.005 | 0.2285 | No | ||
| 147 | TEAD1 | 18121 | 10267 | -0.005 | 0.2221 | No | ||
| 148 | PRDM1 | 19775 3337 | 10270 | -0.005 | 0.2220 | No | ||
| 149 | HOXA10 | 17145 989 | 10319 | -0.005 | 0.2195 | No | ||
| 150 | B3GALT2 | 6498 | 10391 | -0.005 | 0.2157 | No | ||
| 151 | HDAC9 | 21084 2131 | 10394 | -0.005 | 0.2156 | No | ||
| 152 | VAX1 | 23636 | 10658 | -0.006 | 0.2014 | No | ||
| 153 | TENC1 | 22349 9927 | 10661 | -0.006 | 0.2013 | No | ||
| 154 | EIF4E | 15403 1827 8890 | 10775 | -0.006 | 0.1953 | No | ||
| 155 | GREB1 | 6490 2152 | 10947 | -0.007 | 0.1860 | No | ||
| 156 | MFAP5 | 17296 | 11024 | -0.007 | 0.1820 | No | ||
| 157 | SFRP1 | 9805 | 11085 | -0.007 | 0.1788 | No | ||
| 158 | PCDH12 | 7005 | 11221 | -0.008 | 0.1715 | No | ||
| 159 | PCDH18 | 15348 | 11231 | -0.008 | 0.1711 | No | ||
| 160 | NDST4 | 12259 7228 | 11304 | -0.008 | 0.1673 | No | ||
| 161 | ADAMTS5 | 6207 | 11338 | -0.008 | 0.1656 | No | ||
| 162 | DMP1 | 16776 | 11374 | -0.008 | 0.1638 | No | ||
| 163 | FGFR1 | 3789 8968 | 11379 | -0.008 | 0.1637 | No | ||
| 164 | UBE3A | 1209 1084 1830 | 11497 | -0.009 | 0.1574 | No | ||
| 165 | SPAG9 | 1322 12904 7707 | 11532 | -0.009 | 0.1557 | No | ||
| 166 | WEE1 | 18127 | 11768 | -0.010 | 0.1430 | No | ||
| 167 | YWHAZ | 10370 | 11808 | -0.010 | 0.1410 | No | ||
| 168 | MPZ | 9408 | 11859 | -0.010 | 0.1384 | No | ||
| 169 | CNN1 | 8759 | 11905 | -0.011 | 0.1360 | No | ||
| 170 | SPATS2 | 13044 | 12059 | -0.011 | 0.1278 | No | ||
| 171 | TNRC4 | 15514 | 12070 | -0.012 | 0.1274 | No | ||
| 172 | LOXL4 | 23672 | 12077 | -0.012 | 0.1272 | No | ||
| 173 | HAPLN2 | 15296 | 12165 | -0.012 | 0.1226 | No | ||
| 174 | KLHL13 | 12536 | 12221 | -0.012 | 0.1198 | No | ||
| 175 | PAX3 | 13910 | 12302 | -0.013 | 0.1156 | No | ||
| 176 | FOSL2 | 4733 8978 16878 | 12417 | -0.013 | 0.1095 | No | ||
| 177 | KIF1C | 4951 | 12576 | -0.014 | 0.1011 | No | ||
| 178 | SDCCAG8 | 14034 | 12585 | -0.015 | 0.1008 | No | ||
| 179 | MLLT6 | 20678 | 12595 | -0.015 | 0.1005 | No | ||
| 180 | BNC2 | 15850 | 12671 | -0.015 | 0.0966 | No | ||
| 181 | IGF2R | 23123 | 12696 | -0.015 | 0.0955 | No | ||
| 182 | IL17B | 12074 | 12719 | -0.015 | 0.0944 | No | ||
| 183 | TRPM3 | 23904 10357 5927 | 12743 | -0.016 | 0.0934 | No | ||
| 184 | FGFR3 | 8969 3566 | 12834 | -0.016 | 0.0886 | No | ||
| 185 | SEMA6C | 1797 5424 | 12858 | -0.016 | 0.0876 | No | ||
| 186 | EDN1 | 21658 | 12864 | -0.016 | 0.0875 | No | ||
| 187 | MGAT5B | 20584 11197 | 12941 | -0.017 | 0.0836 | No | ||
| 188 | PCSK2 | 9537 5231 14824 | 12993 | -0.017 | 0.0810 | No | ||
| 189 | ETS1 | 10715 6230 3135 | 13041 | -0.018 | 0.0786 | No | ||
| 190 | NAV1 | 3978 5681 | 13148 | -0.019 | 0.0731 | No | ||
| 191 | TNNI1 | 14116 | 13248 | -0.020 | 0.0679 | No | ||
| 192 | FGF10 | 4719 8962 | 13261 | -0.020 | 0.0675 | No | ||
| 193 | ERG | 1686 8915 | 13281 | -0.020 | 0.0667 | No | ||
| 194 | FLI1 | 4729 | 13349 | -0.021 | 0.0633 | No | ||
| 195 | PTPN12 | 16597 3502 3586 3665 | 13458 | -0.022 | 0.0576 | No | ||
| 196 | MID1 | 5097 5098 | 13477 | -0.022 | 0.0569 | No | ||
| 197 | PAQR4 | 23105 | 13521 | -0.023 | 0.0548 | No | ||
| 198 | PLXDC2 | 15114 | 13525 | -0.023 | 0.0549 | No | ||
| 199 | ERH | 9614 4681 | 13528 | -0.023 | 0.0551 | No | ||
| 200 | GJA12 | 20433 1323 | 13609 | -0.024 | 0.0510 | No | ||
| 201 | MDFI | 22950 | 13625 | -0.024 | 0.0505 | No | ||
| 202 | PCBP4 | 12235 | 13808 | -0.027 | 0.0409 | No | ||
| 203 | PANK1 | 1687 23692 | 13833 | -0.028 | 0.0399 | No | ||
| 204 | SLC6A9 | 4783 | 13837 | -0.028 | 0.0400 | No | ||
| 205 | SOX2 | 9849 15612 | 13859 | -0.028 | 0.0392 | No | ||
| 206 | PHF6 | 2648 24340 | 13877 | -0.028 | 0.0386 | No | ||
| 207 | WNT1 | 22371 | 13906 | -0.029 | 0.0374 | No | ||
| 208 | SLC39A8 | 12547 1776 | 14177 | -0.034 | 0.0231 | No | ||
| 209 | ARF4 | 22072 | 14315 | -0.037 | 0.0161 | No | ||
| 210 | FOXP2 | 4312 17523 8523 | 14370 | -0.039 | 0.0136 | No | ||
| 211 | HOXB9 | 20689 | 14402 | -0.039 | 0.0123 | No | ||
| 212 | NDRG2 | 21843 | 14483 | -0.041 | 0.0085 | No | ||
| 213 | SNAPC3 | 13397 2456 8085 | 14540 | -0.043 | 0.0059 | No | ||
| 214 | FGD4 | 10306 5870 1717 1669 1700 | 14625 | -0.046 | 0.0018 | No | ||
| 215 | NUMBL | 1663 9496 | 14676 | -0.047 | -0.0003 | No | ||
| 216 | IGF1 | 3352 9156 3409 | 14702 | -0.048 | -0.0011 | No | ||
| 217 | DLC1 | 11931 3855 | 14705 | -0.048 | -0.0007 | No | ||
| 218 | CDK8 | 6455 6456 3585 3542 | 14717 | -0.049 | -0.0007 | No | ||
| 219 | DPYSL5 | 92 | 15243 | -0.076 | -0.0285 | No | ||
| 220 | DYRK1A | 4649 | 15249 | -0.076 | -0.0278 | No | ||
| 221 | NR2F6 | 4651 8876 18848 8912 | 15272 | -0.078 | -0.0282 | No | ||
| 222 | PAK2 | 10310 5875 | 15410 | -0.087 | -0.0346 | No | ||
| 223 | STMN1 | 9261 | 15569 | -0.101 | -0.0421 | No | ||
| 224 | ILF2 | 12582 | 15580 | -0.102 | -0.0414 | No | ||
| 225 | BCL2L11 | 2790 14861 | 15946 | -0.148 | -0.0596 | No | ||
| 226 | MYST3 | 18651 | 15979 | -0.156 | -0.0596 | No | ||
| 227 | HSPG2 | 4884 16023 | 16031 | -0.164 | -0.0605 | No | ||
| 228 | RDH10 | 8272 | 16056 | -0.167 | -0.0598 | No | ||
| 229 | STAG3 | 16652 | 16123 | -0.182 | -0.0613 | No | ||
| 230 | FEN1 | 8961 | 16172 | -0.189 | -0.0618 | No | ||
| 231 | CRMP1 | 16860 | 16213 | -0.198 | -0.0617 | No | ||
| 232 | BRPF1 | 17332 | 16327 | -0.224 | -0.0653 | No | ||
| 233 | HYAL2 | 4891 | 16376 | -0.236 | -0.0652 | No | ||
| 234 | KPNA3 | 21797 | 16540 | -0.279 | -0.0708 | No | ||
| 235 | RKHD1 | 10693 | 16550 | -0.281 | -0.0681 | No | ||
| 236 | GTF2E2 | 18635 | 16599 | -0.293 | -0.0674 | No | ||
| 237 | PDE7A | 9544 1788 | 16789 | -0.343 | -0.0737 | No | ||
| 238 | ZCCHC8 | 3570 7696 | 16849 | -0.361 | -0.0728 | No | ||
| 239 | CDCA7 | 12437 | 16957 | -0.393 | -0.0741 | No | ||
| 240 | MPHOSPH1 | 23878 | 17060 | -0.432 | -0.0747 | No | ||
| 241 | NXN | 5204 | 17384 | -0.570 | -0.0857 | No | ||
| 242 | YWHAQ | 10369 | 17620 | -0.697 | -0.0905 | No | ||
| 243 | TRIB2 | 21102 | 17771 | -0.796 | -0.0896 | No | ||
| 244 | HTF9C | 4886 1678 | 18012 | -0.992 | -0.0912 | No | ||
| 245 | HMGN1 | 4856 | 18094 | -1.090 | -0.0831 | No | ||
| 246 | NASP | 2383 2387 6955 | 18123 | -1.135 | -0.0716 | No | ||
| 247 | DTX1 | 16396 | 18228 | -1.306 | -0.0623 | No | ||
| 248 | EIF3S10 | 4659 8887 | 18353 | -1.547 | -0.0513 | No | ||
| 249 | PSIP1 | 4159 2507 8319 | 18376 | -1.606 | -0.0341 | No | ||
| 250 | TPM3 | 12233 7209 7208 1790 | 18397 | -1.665 | -0.0160 | No | ||
| 251 | SCD | 23662 | 18531 | -2.432 | 0.0046 | No |