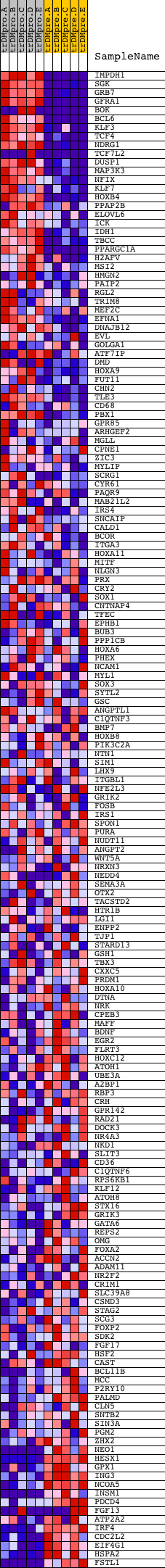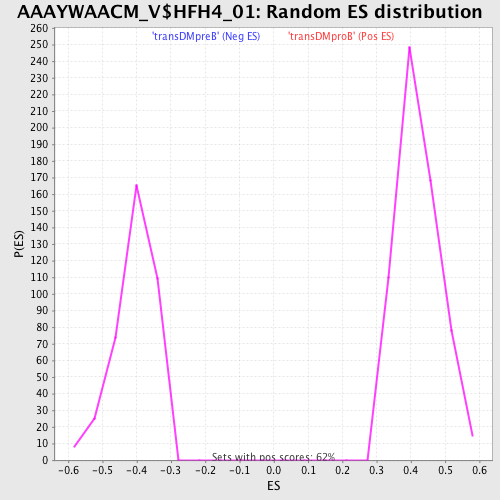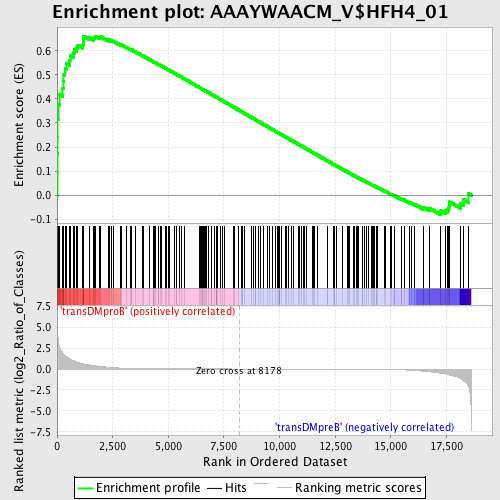
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | AAAYWAACM_V$HFH4_01 |
| Enrichment Score (ES) | 0.66108996 |
| Normalized Enrichment Score (NES) | 1.569817 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.058624666 |
| FWER p-Value | 0.391 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IMPDH1 | 17197 1131 | 9 | 5.245 | 0.0895 | Yes | ||
| 2 | SGK | 9809 3419 | 12 | 4.998 | 0.1750 | Yes | ||
| 3 | GRB7 | 20673 | 25 | 4.169 | 0.2459 | Yes | ||
| 4 | GFRA1 | 9015 | 28 | 4.130 | 0.3166 | Yes | ||
| 5 | BOK | 11953 14174 | 44 | 3.650 | 0.3784 | Yes | ||
| 6 | BCL6 | 22624 | 112 | 2.561 | 0.4187 | Yes | ||
| 7 | KLF3 | 4961 | 257 | 1.890 | 0.4433 | Yes | ||
| 8 | TCF4 | 5642 | 292 | 1.758 | 0.4716 | Yes | ||
| 9 | NDRG1 | 22276 | 305 | 1.727 | 0.5005 | Yes | ||
| 10 | TCF7L2 | 10048 5646 | 354 | 1.605 | 0.5254 | Yes | ||
| 11 | DUSP1 | 23061 | 416 | 1.523 | 0.5482 | Yes | ||
| 12 | MAP3K3 | 20626 | 561 | 1.226 | 0.5615 | Yes | ||
| 13 | NFIX | 9458 | 582 | 1.194 | 0.5808 | Yes | ||
| 14 | KLF7 | 8205 | 734 | 0.981 | 0.5895 | Yes | ||
| 15 | HOXB4 | 20686 | 767 | 0.952 | 0.6041 | Yes | ||
| 16 | PPAP2B | 7503 16161 | 891 | 0.836 | 0.6117 | Yes | ||
| 17 | ELOVL6 | 5009 | 918 | 0.805 | 0.6241 | Yes | ||
| 18 | ICK | 7135 12146 | 1150 | 0.618 | 0.6222 | Yes | ||
| 19 | IDH1 | 4894 | 1164 | 0.610 | 0.6320 | Yes | ||
| 20 | TBCC | 23215 | 1182 | 0.601 | 0.6413 | Yes | ||
| 21 | PPARGC1A | 16533 | 1193 | 0.597 | 0.6510 | Yes | ||
| 22 | H2AFV | 1273 20531 | 1196 | 0.593 | 0.6611 | Yes | ||
| 23 | MSI2 | 8030 1268 | 1451 | 0.482 | 0.6556 | No | ||
| 24 | HMGN2 | 9095 | 1653 | 0.396 | 0.6515 | No | ||
| 25 | PAIP2 | 12593 | 1668 | 0.389 | 0.6574 | No | ||
| 26 | RGL2 | 23289 | 1727 | 0.369 | 0.6606 | No | ||
| 27 | TRIM8 | 23824 | 1921 | 0.310 | 0.6554 | No | ||
| 28 | MEF2C | 3204 9378 | 1945 | 0.302 | 0.6594 | No | ||
| 29 | EFNA1 | 15279 | 2291 | 0.208 | 0.6442 | No | ||
| 30 | DNAJB12 | 7146 | 2333 | 0.196 | 0.6454 | No | ||
| 31 | EVL | 21156 | 2435 | 0.174 | 0.6429 | No | ||
| 32 | GOLGA1 | 14595 | 2532 | 0.156 | 0.6403 | No | ||
| 33 | ATF7IP | 12028 13038 | 2835 | 0.108 | 0.6258 | No | ||
| 34 | DMD | 24295 2647 | 2886 | 0.102 | 0.6249 | No | ||
| 35 | HOXA9 | 17147 9109 1024 | 3098 | 0.081 | 0.6148 | No | ||
| 36 | FUT11 | 13084 | 3277 | 0.065 | 0.6063 | No | ||
| 37 | CHN2 | 7652 | 3342 | 0.060 | 0.6038 | No | ||
| 38 | TLE3 | 5767 19416 | 3343 | 0.060 | 0.6049 | No | ||
| 39 | CD68 | 4501 | 3523 | 0.048 | 0.5960 | No | ||
| 40 | PBX1 | 5224 | 3815 | 0.036 | 0.5808 | No | ||
| 41 | GPR85 | 7227 1048 | 3865 | 0.034 | 0.5787 | No | ||
| 42 | ARHGEF2 | 4987 | 4150 | 0.024 | 0.5638 | No | ||
| 43 | MGLL | 17368 1145 | 4345 | 0.020 | 0.5536 | No | ||
| 44 | CPNE1 | 11186 | 4352 | 0.020 | 0.5536 | No | ||
| 45 | ZIC3 | 10430 | 4386 | 0.019 | 0.5521 | No | ||
| 46 | MYLIP | 21652 3189 | 4439 | 0.018 | 0.5496 | No | ||
| 47 | SCRG1 | 18614 | 4542 | 0.017 | 0.5444 | No | ||
| 48 | CYR61 | 9159 15145 9311 5024 | 4566 | 0.016 | 0.5434 | No | ||
| 49 | PAQR9 | 19351 | 4633 | 0.015 | 0.5401 | No | ||
| 50 | MAB21L2 | 15305 | 4711 | 0.014 | 0.5362 | No | ||
| 51 | IRS4 | 9183 4926 | 4849 | 0.013 | 0.5290 | No | ||
| 52 | SNCAIP | 12589 | 4916 | 0.012 | 0.5256 | No | ||
| 53 | CALD1 | 4273 8463 | 5005 | 0.012 | 0.5210 | No | ||
| 54 | BCOR | 12934 2561 | 5047 | 0.011 | 0.5190 | No | ||
| 55 | ITGA3 | 20284 | 5278 | 0.010 | 0.5067 | No | ||
| 56 | HOXA11 | 17146 | 5351 | 0.009 | 0.5030 | No | ||
| 57 | MITF | 17349 | 5365 | 0.009 | 0.5024 | No | ||
| 58 | NLGN3 | 10878 | 5490 | 0.009 | 0.4959 | No | ||
| 59 | PRX | 3729 9628 | 5578 | 0.008 | 0.4913 | No | ||
| 60 | CRY2 | 14518 | 5703 | 0.008 | 0.4847 | No | ||
| 61 | SOX1 | 18685 | 6403 | 0.005 | 0.4469 | No | ||
| 62 | CNTNAP4 | 18462 3857 | 6444 | 0.005 | 0.4448 | No | ||
| 63 | TFEC | 17213 | 6489 | 0.005 | 0.4425 | No | ||
| 64 | EPHB1 | 19024 | 6532 | 0.004 | 0.4403 | No | ||
| 65 | BUB3 | 18045 | 6540 | 0.004 | 0.4400 | No | ||
| 66 | PPP1CB | 5282 3529 3504 | 6599 | 0.004 | 0.4369 | No | ||
| 67 | HOXA6 | 17150 | 6640 | 0.004 | 0.4348 | No | ||
| 68 | PHEX | 24024 | 6685 | 0.004 | 0.4325 | No | ||
| 69 | NCAM1 | 5149 | 6694 | 0.004 | 0.4321 | No | ||
| 70 | MYL1 | 4015 | 6797 | 0.004 | 0.4266 | No | ||
| 71 | SOX3 | 24145 | 6921 | 0.003 | 0.4200 | No | ||
| 72 | SYTL2 | 18190 1539 3730 | 6956 | 0.003 | 0.4182 | No | ||
| 73 | GSC | 20990 | 7090 | 0.003 | 0.4111 | No | ||
| 74 | ANGPTL1 | 10195 8589 13064 | 7166 | 0.003 | 0.4071 | No | ||
| 75 | C1QTNF3 | 2224 22505 | 7174 | 0.003 | 0.4067 | No | ||
| 76 | BMP7 | 14324 | 7178 | 0.003 | 0.4066 | No | ||
| 77 | HOXB8 | 9111 | 7206 | 0.003 | 0.4052 | No | ||
| 78 | PIK3C2A | 17667 | 7330 | 0.002 | 0.3986 | No | ||
| 79 | NTN1 | 20400 | 7415 | 0.002 | 0.3940 | No | ||
| 80 | SIM1 | 20031 | 7542 | 0.002 | 0.3872 | No | ||
| 81 | LHX9 | 9276 | 7920 | 0.001 | 0.3668 | No | ||
| 82 | ITGBL1 | 5850 | 7992 | 0.000 | 0.3630 | No | ||
| 83 | NFE2L3 | 17446 | 8172 | 0.000 | 0.3533 | No | ||
| 84 | GRIK2 | 3318 | 8286 | -0.000 | 0.3471 | No | ||
| 85 | FOSB | 17945 | 8326 | -0.000 | 0.3450 | No | ||
| 86 | IRS1 | 4925 | 8350 | -0.000 | 0.3438 | No | ||
| 87 | SPON1 | 10600 | 8432 | -0.001 | 0.3394 | No | ||
| 88 | PURA | 9670 | 8758 | -0.001 | 0.3218 | No | ||
| 89 | NUDT11 | 24387 | 8806 | -0.001 | 0.3193 | No | ||
| 90 | ANGPT2 | 14885 3783 18923 | 8911 | -0.002 | 0.3137 | No | ||
| 91 | WNT5A | 22066 5880 | 8917 | -0.002 | 0.3134 | No | ||
| 92 | NRXN3 | 2153 5196 | 9052 | -0.002 | 0.3062 | No | ||
| 93 | NEDD4 | 19384 3101 | 9119 | -0.002 | 0.3027 | No | ||
| 94 | SEMA3A | 16919 | 9278 | -0.003 | 0.2941 | No | ||
| 95 | OTX2 | 9519 | 9449 | -0.003 | 0.2850 | No | ||
| 96 | TACSTD2 | 17126 | 9532 | -0.003 | 0.2806 | No | ||
| 97 | HTR1B | 19051 | 9669 | -0.003 | 0.2733 | No | ||
| 98 | LGI1 | 23867 | 9798 | -0.004 | 0.2664 | No | ||
| 99 | ENPP2 | 9548 | 9926 | -0.004 | 0.2596 | No | ||
| 100 | TJP1 | 17803 | 9967 | -0.004 | 0.2575 | No | ||
| 101 | STARD13 | 10833 | 9995 | -0.004 | 0.2561 | No | ||
| 102 | GSH1 | 16622 | 10004 | -0.004 | 0.2557 | No | ||
| 103 | TBX3 | 16723 | 10008 | -0.004 | 0.2556 | No | ||
| 104 | CXXC5 | 12523 | 10097 | -0.004 | 0.2509 | No | ||
| 105 | PRDM1 | 19775 3337 | 10270 | -0.005 | 0.2417 | No | ||
| 106 | HOXA10 | 17145 989 | 10319 | -0.005 | 0.2392 | No | ||
| 107 | DTNA | 8870 4647 23611 1991 | 10399 | -0.005 | 0.2350 | No | ||
| 108 | NRK | 6559 | 10542 | -0.006 | 0.2274 | No | ||
| 109 | CPEB3 | 5539 | 10620 | -0.006 | 0.2233 | No | ||
| 110 | MAFF | 22423 2243 | 10839 | -0.007 | 0.2116 | No | ||
| 111 | BDNF | 14926 2797 | 10903 | -0.007 | 0.2083 | No | ||
| 112 | EGR2 | 8886 | 10977 | -0.007 | 0.2045 | No | ||
| 113 | FLRT3 | 7730 12930 2673 | 11063 | -0.007 | 0.2000 | No | ||
| 114 | HOXC12 | 22343 | 11134 | -0.008 | 0.1963 | No | ||
| 115 | ATOH1 | 4419 | 11187 | -0.008 | 0.1937 | No | ||
| 116 | UBE3A | 1209 1084 1830 | 11497 | -0.009 | 0.1771 | No | ||
| 117 | A2BP1 | 11212 1652 1689 | 11544 | -0.009 | 0.1747 | No | ||
| 118 | RBP3 | 22047 | 11559 | -0.009 | 0.1741 | No | ||
| 119 | CRH | 8784 | 11723 | -0.010 | 0.1654 | No | ||
| 120 | GPR142 | 20604 | 12148 | -0.012 | 0.1427 | No | ||
| 121 | RAD21 | 22298 | 12173 | -0.012 | 0.1416 | No | ||
| 122 | DOCK3 | 9920 5537 10751 | 12401 | -0.013 | 0.1295 | No | ||
| 123 | NR4A3 | 9473 16212 5183 | 12466 | -0.014 | 0.1263 | No | ||
| 124 | NKD1 | 8228 | 12481 | -0.014 | 0.1257 | No | ||
| 125 | SLIT3 | 20925 | 12553 | -0.014 | 0.1221 | No | ||
| 126 | CD36 | 8712 | 12827 | -0.016 | 0.1076 | No | ||
| 127 | C1QTNF6 | 13063 | 13036 | -0.018 | 0.0966 | No | ||
| 128 | RPS6KB1 | 7815 1207 13040 | 13079 | -0.018 | 0.0947 | No | ||
| 129 | KLF12 | 4960 2637 21732 9228 | 13156 | -0.019 | 0.0909 | No | ||
| 130 | ATOH8 | 17120 | 13321 | -0.020 | 0.0823 | No | ||
| 131 | STX16 | 10464 | 13372 | -0.021 | 0.0800 | No | ||
| 132 | GRIK3 | 16083 | 13472 | -0.022 | 0.0750 | No | ||
| 133 | GATA6 | 96 | 13511 | -0.023 | 0.0733 | No | ||
| 134 | REPS2 | 24018 | 13554 | -0.023 | 0.0714 | No | ||
| 135 | OMG | 5209 | 13730 | -0.026 | 0.0624 | No | ||
| 136 | FOXA2 | 14405 2889 | 13734 | -0.026 | 0.0627 | No | ||
| 137 | ACCN2 | 22363 | 13812 | -0.027 | 0.0590 | No | ||
| 138 | ADAM11 | 4340 1432 | 13923 | -0.029 | 0.0535 | No | ||
| 139 | NR2F2 | 8619 409 | 14008 | -0.031 | 0.0495 | No | ||
| 140 | CRIM1 | 403 | 14139 | -0.033 | 0.0430 | No | ||
| 141 | SLC39A8 | 12547 1776 | 14177 | -0.034 | 0.0416 | No | ||
| 142 | CSMD3 | 10735 7938 | 14203 | -0.034 | 0.0408 | No | ||
| 143 | STAG2 | 5521 | 14273 | -0.036 | 0.0377 | No | ||
| 144 | SCG3 | 19061 | 14367 | -0.038 | 0.0333 | No | ||
| 145 | FOXP2 | 4312 17523 8523 | 14370 | -0.039 | 0.0338 | No | ||
| 146 | SDK2 | 6208 20162 | 14416 | -0.040 | 0.0321 | No | ||
| 147 | FGF17 | 21757 | 14703 | -0.048 | 0.0174 | No | ||
| 148 | HSF2 | 9128 | 14760 | -0.050 | 0.0152 | No | ||
| 149 | CAST | 3182 4478 3177 | 15001 | -0.061 | 0.0033 | No | ||
| 150 | BCL11B | 7190 | 15046 | -0.064 | 0.0020 | No | ||
| 151 | MCC | 552 | 15167 | -0.071 | -0.0033 | No | ||
| 152 | P2RY10 | 2613 24269 | 15183 | -0.072 | -0.0029 | No | ||
| 153 | PALMD | 15181 | 15496 | -0.094 | -0.0182 | No | ||
| 154 | CLN5 | 21943 | 15499 | -0.094 | -0.0167 | No | ||
| 155 | SNTB2 | 3887 18476 | 15594 | -0.103 | -0.0200 | No | ||
| 156 | SIN3A | 5442 | 15834 | -0.133 | -0.0307 | No | ||
| 157 | PGM2 | 16168 | 15929 | -0.146 | -0.0333 | No | ||
| 158 | ZHX2 | 11842 | 16078 | -0.171 | -0.0384 | No | ||
| 159 | NEO1 | 5161 | 16447 | -0.254 | -0.0540 | No | ||
| 160 | HESX1 | 22070 | 16470 | -0.259 | -0.0507 | No | ||
| 161 | GPX1 | 19310 | 16720 | -0.324 | -0.0587 | No | ||
| 162 | ING3 | 17514 1019 | 16747 | -0.331 | -0.0544 | No | ||
| 163 | NCOA5 | 14341 | 17226 | -0.490 | -0.0719 | No | ||
| 164 | INSM1 | 14815 | 17253 | -0.507 | -0.0646 | No | ||
| 165 | PDCD4 | 5232 23816 | 17458 | -0.603 | -0.0653 | No | ||
| 166 | FGF13 | 4720 8963 2627 | 17551 | -0.656 | -0.0591 | No | ||
| 167 | ATP2A2 | 4421 3481 | 17610 | -0.694 | -0.0503 | No | ||
| 168 | IRF4 | 21679 | 17618 | -0.695 | -0.0388 | No | ||
| 169 | CDC2L2 | 15964 2419 | 17650 | -0.712 | -0.0283 | No | ||
| 170 | EIF4G1 | 22818 | 18146 | -1.151 | -0.0354 | No | ||
| 171 | HSPA2 | 21237 | 18261 | -1.355 | -0.0183 | No | ||
| 172 | FSTL1 | 22765 | 18495 | -2.186 | 0.0066 | No |