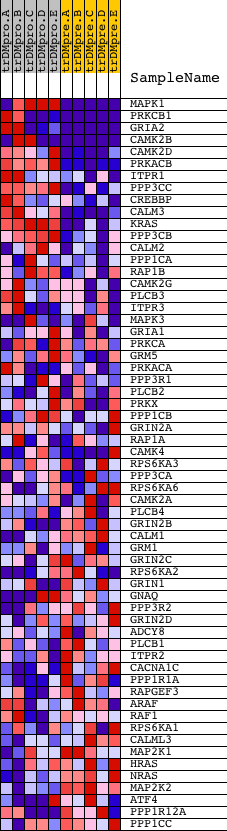
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
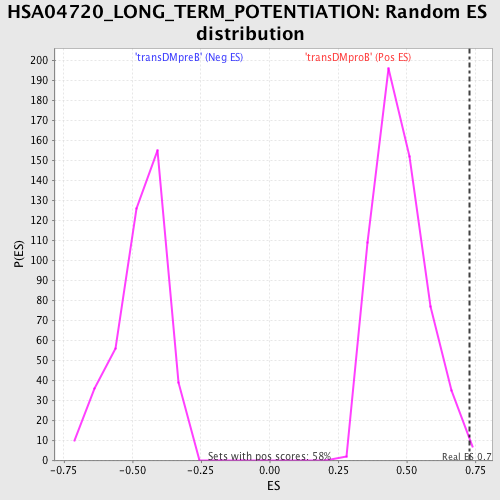
| GeneSet | HSA04720_LONG_TERM_POTENTIATION |
| Enrichment Score (ES) | 0.73029846 |
| Normalized Enrichment Score (NES) | 1.5327852 |
| Nominal p-value | 0.0051903115 |
| FDR q-value | 0.2600549 |
| FWER p-Value | 0.659 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAPK1 | 1642 11167 | 80 | 2.911 | 0.1509 | Yes | ||
| 2 | PRKCB1 | 1693 9574 | 81 | 2.858 | 0.3034 | Yes | ||
| 3 | GRIA2 | 4802 | 295 | 1.754 | 0.3854 | Yes | ||
| 4 | CAMK2B | 20536 | 391 | 1.552 | 0.4631 | Yes | ||
| 5 | CAMK2D | 4232 | 680 | 1.045 | 0.5033 | Yes | ||
| 6 | PRKACB | 15140 | 705 | 1.014 | 0.5561 | Yes | ||
| 7 | ITPR1 | 17341 | 884 | 0.841 | 0.5913 | Yes | ||
| 8 | PPP3CC | 21763 | 1251 | 0.569 | 0.6020 | Yes | ||
| 9 | CREBBP | 22682 8783 | 1308 | 0.542 | 0.6279 | Yes | ||
| 10 | CALM3 | 8682 | 1340 | 0.528 | 0.6544 | Yes | ||
| 11 | KRAS | 9247 | 1356 | 0.521 | 0.6813 | Yes | ||
| 12 | PPP3CB | 5285 | 1499 | 0.461 | 0.6983 | Yes | ||
| 13 | CALM2 | 8681 | 1771 | 0.355 | 0.7026 | Yes | ||
| 14 | PPP1CA | 9608 3756 914 909 | 1801 | 0.344 | 0.7194 | Yes | ||
| 15 | RAP1B | 10083 | 1909 | 0.312 | 0.7303 | Yes | ||
| 16 | CAMK2G | 21905 | 2283 | 0.210 | 0.7214 | No | ||
| 17 | PLCB3 | 23799 | 2591 | 0.147 | 0.7127 | No | ||
| 18 | ITPR3 | 9195 | 3225 | 0.069 | 0.6823 | No | ||
| 19 | MAPK3 | 6458 11170 | 4552 | 0.016 | 0.6117 | No | ||
| 20 | GRIA1 | 20877 4801 9040 | 4640 | 0.015 | 0.6078 | No | ||
| 21 | PRKCA | 20174 | 4949 | 0.012 | 0.5919 | No | ||
| 22 | GRM5 | 8426 | 5338 | 0.010 | 0.5715 | No | ||
| 23 | PRKACA | 18549 3844 | 5640 | 0.008 | 0.5557 | No | ||
| 24 | PPP3R1 | 9611 489 | 5824 | 0.007 | 0.5462 | No | ||
| 25 | PLCB2 | 5262 | 6175 | 0.006 | 0.5276 | No | ||
| 26 | PRKX | 9619 5290 | 6327 | 0.005 | 0.5197 | No | ||
| 27 | PPP1CB | 5282 3529 3504 | 6599 | 0.004 | 0.5054 | No | ||
| 28 | GRIN2A | 22668 | 7235 | 0.002 | 0.4713 | No | ||
| 29 | RAP1A | 8467 | 8098 | 0.000 | 0.4248 | No | ||
| 30 | CAMK4 | 4473 | 8169 | 0.000 | 0.4211 | No | ||
| 31 | RPS6KA3 | 8490 | 9927 | -0.004 | 0.3266 | No | ||
| 32 | PPP3CA | 1863 5284 | 10074 | -0.004 | 0.3190 | No | ||
| 33 | RPS6KA6 | 12458 | 10187 | -0.005 | 0.3132 | No | ||
| 34 | CAMK2A | 2024 23541 1980 | 10341 | -0.005 | 0.3052 | No | ||
| 35 | PLCB4 | 9586 | 10847 | -0.007 | 0.2783 | No | ||
| 36 | GRIN2B | 16957 | 11216 | -0.008 | 0.2589 | No | ||
| 37 | CALM1 | 21184 | 11378 | -0.008 | 0.2507 | No | ||
| 38 | GRM1 | 19822 | 11433 | -0.009 | 0.2483 | No | ||
| 39 | GRIN2C | 20155 | 11833 | -0.010 | 0.2273 | No | ||
| 40 | RPS6KA2 | 9759 9758 | 11842 | -0.010 | 0.2274 | No | ||
| 41 | GRIN1 | 9041 4804 | 12334 | -0.013 | 0.2017 | No | ||
| 42 | GNAQ | 4786 23909 3685 | 12410 | -0.013 | 0.1983 | No | ||
| 43 | PPP3R2 | 9612 | 12427 | -0.013 | 0.1982 | No | ||
| 44 | GRIN2D | 9042 | 12722 | -0.015 | 0.1832 | No | ||
| 45 | ADCY8 | 22281 | 12754 | -0.016 | 0.1823 | No | ||
| 46 | PLCB1 | 14832 2821 | 13146 | -0.019 | 0.1623 | No | ||
| 47 | ITPR2 | 9194 | 13771 | -0.026 | 0.1300 | No | ||
| 48 | CACNA1C | 1158 1166 24464 4463 8674 | 13840 | -0.028 | 0.1278 | No | ||
| 49 | PPP1R1A | 22102 | 14164 | -0.034 | 0.1122 | No | ||
| 50 | RAPGEF3 | 22146 | 14372 | -0.039 | 0.1031 | No | ||
| 51 | ARAF | 24367 | 14892 | -0.055 | 0.0781 | No | ||
| 52 | RAF1 | 17035 | 15064 | -0.065 | 0.0724 | No | ||
| 53 | RPS6KA1 | 15725 | 15263 | -0.077 | 0.0658 | No | ||
| 54 | CALML3 | 21553 | 15797 | -0.128 | 0.0439 | No | ||
| 55 | MAP2K1 | 19082 | 15878 | -0.140 | 0.0470 | No | ||
| 56 | HRAS | 4868 | 16140 | -0.184 | 0.0428 | No | ||
| 57 | NRAS | 5191 | 16367 | -0.233 | 0.0430 | No | ||
| 58 | MAP2K2 | 19933 | 16484 | -0.264 | 0.0509 | No | ||
| 59 | ATF4 | 22417 2239 | 16600 | -0.293 | 0.0603 | No | ||
| 60 | PPP1R12A | 3354 19886 | 16837 | -0.357 | 0.0666 | No | ||
| 61 | PPP1CC | 9609 5283 | 17345 | -0.548 | 0.0685 | No |