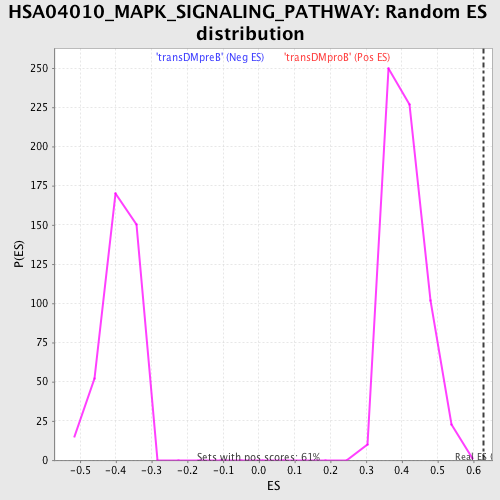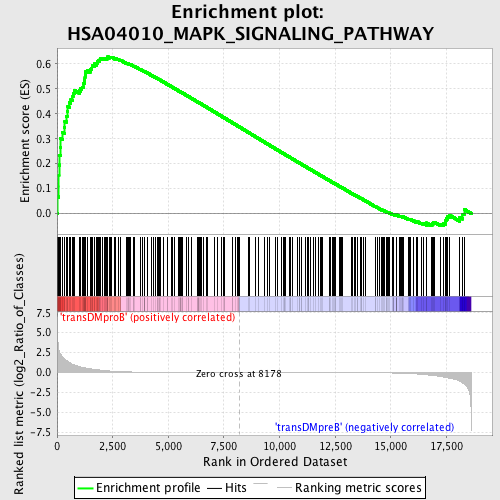
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | HSA04010_MAPK_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.6277755 |
| Normalized Enrichment Score (NES) | 1.537198 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.297479 |
| FWER p-Value | 0.625 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DUSP6 | 19891 3399 | 15 | 4.550 | 0.0684 | Yes | ||
| 2 | MAPK1 | 1642 11167 | 80 | 2.911 | 0.1092 | Yes | ||
| 3 | PRKCB1 | 1693 9574 | 81 | 2.858 | 0.1526 | Yes | ||
| 4 | MAPK9 | 1233 20903 1383 | 85 | 2.800 | 0.1950 | Yes | ||
| 5 | RRAS2 | 12431 | 110 | 2.587 | 0.2331 | Yes | ||
| 6 | MAPKAPK2 | 13838 | 146 | 2.333 | 0.2666 | Yes | ||
| 7 | CACNA2D1 | 8676 | 157 | 2.291 | 0.3009 | Yes | ||
| 8 | PPM1B | 5280 1567 5281 | 240 | 1.952 | 0.3262 | Yes | ||
| 9 | RASGRP2 | 9693 | 332 | 1.658 | 0.3464 | Yes | ||
| 10 | MAP4K2 | 6457 | 348 | 1.618 | 0.3702 | Yes | ||
| 11 | DUSP1 | 23061 | 416 | 1.523 | 0.3897 | Yes | ||
| 12 | TGFB1 | 18332 | 452 | 1.452 | 0.4099 | Yes | ||
| 13 | GADD45A | 17129 | 481 | 1.383 | 0.4294 | Yes | ||
| 14 | MAP3K3 | 20626 | 561 | 1.226 | 0.4437 | Yes | ||
| 15 | DUSP2 | 14865 | 619 | 1.119 | 0.4577 | Yes | ||
| 16 | PRKACB | 15140 | 705 | 1.014 | 0.4685 | Yes | ||
| 17 | DDIT3 | 19857 | 721 | 0.999 | 0.4828 | Yes | ||
| 18 | RASA1 | 10174 | 782 | 0.936 | 0.4938 | Yes | ||
| 19 | MAP3K5 | 20079 11166 | 996 | 0.734 | 0.4934 | Yes | ||
| 20 | TRAF6 | 5797 14940 | 1054 | 0.682 | 0.5006 | Yes | ||
| 21 | MAP4K3 | 22887 | 1118 | 0.641 | 0.5070 | Yes | ||
| 22 | MAPK14 | 23313 | 1184 | 0.600 | 0.5125 | Yes | ||
| 23 | MAP3K7IP2 | 19827 | 1186 | 0.599 | 0.5216 | Yes | ||
| 24 | NFKB2 | 23810 | 1233 | 0.578 | 0.5279 | Yes | ||
| 25 | CRKL | 4560 | 1245 | 0.572 | 0.5360 | Yes | ||
| 26 | PPP3CC | 21763 | 1251 | 0.569 | 0.5444 | Yes | ||
| 27 | RAC2 | 22217 | 1257 | 0.566 | 0.5527 | Yes | ||
| 28 | AKT3 | 13739 982 | 1267 | 0.562 | 0.5608 | Yes | ||
| 29 | MAP3K14 | 11998 | 1289 | 0.549 | 0.5680 | Yes | ||
| 30 | KRAS | 9247 | 1356 | 0.521 | 0.5723 | Yes | ||
| 31 | PTPN7 | 11500 | 1482 | 0.468 | 0.5726 | Yes | ||
| 32 | PPP3CB | 5285 | 1499 | 0.461 | 0.5788 | Yes | ||
| 33 | RRAS | 18256 | 1543 | 0.442 | 0.5831 | Yes | ||
| 34 | RAC1 | 16302 | 1568 | 0.428 | 0.5883 | Yes | ||
| 35 | JUN | 15832 | 1569 | 0.428 | 0.5948 | Yes | ||
| 36 | MAP4K4 | 11241 | 1693 | 0.381 | 0.5940 | Yes | ||
| 37 | NFKB1 | 15160 | 1695 | 0.381 | 0.5997 | Yes | ||
| 38 | MKNK2 | 3299 9392 | 1789 | 0.349 | 0.5999 | Yes | ||
| 39 | RPS6KA4 | 23804 | 1799 | 0.345 | 0.6047 | Yes | ||
| 40 | CASP3 | 8693 | 1817 | 0.337 | 0.6089 | Yes | ||
| 41 | STK3 | 7084 | 1848 | 0.328 | 0.6122 | Yes | ||
| 42 | RAP1B | 10083 | 1909 | 0.312 | 0.6137 | Yes | ||
| 43 | STK4 | 14743 | 1926 | 0.308 | 0.6176 | Yes | ||
| 44 | MEF2C | 3204 9378 | 1945 | 0.302 | 0.6212 | Yes | ||
| 45 | CDC25B | 14841 | 2033 | 0.276 | 0.6206 | Yes | ||
| 46 | ARRB2 | 20806 | 2116 | 0.251 | 0.6200 | Yes | ||
| 47 | MAP3K1 | 21348 | 2190 | 0.232 | 0.6195 | Yes | ||
| 48 | MAP2K3 | 20856 | 2228 | 0.221 | 0.6209 | Yes | ||
| 49 | SOS2 | 21049 | 2262 | 0.214 | 0.6223 | Yes | ||
| 50 | MAP2K5 | 19088 | 2276 | 0.211 | 0.6249 | Yes | ||
| 51 | MAP3K4 | 23126 | 2282 | 0.210 | 0.6278 | Yes | ||
| 52 | IL1B | 14431 | 2372 | 0.186 | 0.6258 | No | ||
| 53 | CACNB2 | 4466 8678 | 2406 | 0.180 | 0.6267 | No | ||
| 54 | GNG12 | 4789 | 2454 | 0.170 | 0.6267 | No | ||
| 55 | ACVR1B | 4335 22353 | 2599 | 0.145 | 0.6211 | No | ||
| 56 | EGF | 15169 | 2619 | 0.142 | 0.6222 | No | ||
| 57 | GRB2 | 20149 | 2736 | 0.123 | 0.6178 | No | ||
| 58 | CD14 | 23452 | 2841 | 0.108 | 0.6138 | No | ||
| 59 | PAK1 | 9527 | 2868 | 0.104 | 0.6139 | No | ||
| 60 | RASGRP4 | 6119 | 3132 | 0.078 | 0.6008 | No | ||
| 61 | MYC | 22465 9435 | 3151 | 0.076 | 0.6010 | No | ||
| 62 | FGFR2 | 1917 4722 1119 | 3186 | 0.073 | 0.6003 | No | ||
| 63 | MAPK13 | 23314 | 3196 | 0.072 | 0.6009 | No | ||
| 64 | FOS | 21202 | 3267 | 0.066 | 0.5981 | No | ||
| 65 | GNA12 | 9023 | 3320 | 0.062 | 0.5962 | No | ||
| 66 | NF1 | 5165 | 3443 | 0.054 | 0.5903 | No | ||
| 67 | ELK4 | 8895 4664 3937 | 3449 | 0.053 | 0.5909 | No | ||
| 68 | PPM1A | 5279 9607 | 3495 | 0.050 | 0.5892 | No | ||
| 69 | CACNA1F | 12054 24393 | 3737 | 0.038 | 0.5766 | No | ||
| 70 | FAS | 23881 3750 | 3746 | 0.038 | 0.5768 | No | ||
| 71 | PDGFRB | 23539 | 3843 | 0.035 | 0.5721 | No | ||
| 72 | CACNA2D2 | 7154 | 3912 | 0.032 | 0.5689 | No | ||
| 73 | ARRB1 | 3890 8465 18178 4276 | 3916 | 0.032 | 0.5692 | No | ||
| 74 | PDGFA | 5237 | 3931 | 0.031 | 0.5689 | No | ||
| 75 | DUSP4 | 18632 3820 | 3938 | 0.031 | 0.5690 | No | ||
| 76 | FGF1 | 1994 23447 | 4054 | 0.027 | 0.5632 | No | ||
| 77 | MAP3K7IP1 | 2193 2171 22419 | 4071 | 0.027 | 0.5627 | No | ||
| 78 | MAP3K8 | 23495 | 4233 | 0.022 | 0.5543 | No | ||
| 79 | TRAF2 | 14657 | 4330 | 0.020 | 0.5494 | No | ||
| 80 | PLA2G5 | 9583 | 4416 | 0.019 | 0.5451 | No | ||
| 81 | PLA2G1B | 9581 | 4509 | 0.017 | 0.5403 | No | ||
| 82 | FGF22 | 12468 7385 | 4521 | 0.017 | 0.5400 | No | ||
| 83 | MAPK3 | 6458 11170 | 4552 | 0.016 | 0.5386 | No | ||
| 84 | TNF | 23004 | 4599 | 0.016 | 0.5363 | No | ||
| 85 | CACNA1E | 4465 | 4619 | 0.016 | 0.5355 | No | ||
| 86 | MAPK7 | 1381 20414 | 4636 | 0.015 | 0.5349 | No | ||
| 87 | TGFBR1 | 16213 10165 5745 | 4801 | 0.013 | 0.5262 | No | ||
| 88 | PRKCA | 20174 | 4949 | 0.012 | 0.5184 | No | ||
| 89 | DUSP7 | 10661 | 4966 | 0.012 | 0.5177 | No | ||
| 90 | CRK | 4559 1249 | 5134 | 0.011 | 0.5087 | No | ||
| 91 | MAP3K6 | 16054 | 5180 | 0.011 | 0.5065 | No | ||
| 92 | MAP3K10 | 3831 17916 | 5288 | 0.010 | 0.5008 | No | ||
| 93 | FGF21 | 17830 | 5463 | 0.009 | 0.4915 | No | ||
| 94 | FGF9 | 8966 | 5506 | 0.009 | 0.4893 | No | ||
| 95 | DUSP10 | 4003 14016 | 5533 | 0.008 | 0.4880 | No | ||
| 96 | FGFR4 | 3226 | 5546 | 0.008 | 0.4875 | No | ||
| 97 | IL1R2 | 14265 | 5549 | 0.008 | 0.4875 | No | ||
| 98 | CACNA1G | 1237 940 20292 | 5607 | 0.008 | 0.4845 | No | ||
| 99 | PRKACA | 18549 3844 | 5640 | 0.008 | 0.4829 | No | ||
| 100 | SOS1 | 5476 | 5657 | 0.008 | 0.4822 | No | ||
| 101 | PPP3R1 | 9611 489 | 5824 | 0.007 | 0.4732 | No | ||
| 102 | MAP3K13 | 22814 | 5891 | 0.007 | 0.4697 | No | ||
| 103 | FGF18 | 4721 1351 | 5909 | 0.007 | 0.4689 | No | ||
| 104 | DUSP16 | 1004 7699 | 6041 | 0.006 | 0.4619 | No | ||
| 105 | FGF4 | 8964 | 6302 | 0.005 | 0.4478 | No | ||
| 106 | PRKX | 9619 5290 | 6327 | 0.005 | 0.4466 | No | ||
| 107 | CACNG7 | 13497 | 6345 | 0.005 | 0.4457 | No | ||
| 108 | ACVR1C | 14585 | 6349 | 0.005 | 0.4456 | No | ||
| 109 | PLA2G10 | 22658 | 6363 | 0.005 | 0.4450 | No | ||
| 110 | RASGRP1 | 5360 14476 9694 | 6394 | 0.005 | 0.4435 | No | ||
| 111 | TGFBR2 | 2994 3001 10167 | 6464 | 0.005 | 0.4398 | No | ||
| 112 | PDGFB | 22199 2311 | 6497 | 0.005 | 0.4381 | No | ||
| 113 | FGF16 | 24270 | 6572 | 0.004 | 0.4341 | No | ||
| 114 | TAOK2 | 10610 3794 | 6706 | 0.004 | 0.4270 | No | ||
| 115 | CDC42 | 4503 8722 4504 2465 | 6780 | 0.004 | 0.4230 | No | ||
| 116 | PLA2G12B | 20016 | 7070 | 0.003 | 0.4074 | No | ||
| 117 | DUSP8 | 9493 | 7186 | 0.003 | 0.4011 | No | ||
| 118 | MAPT | 9420 1347 | 7406 | 0.002 | 0.3893 | No | ||
| 119 | CACNB4 | 4467 | 7472 | 0.002 | 0.3857 | No | ||
| 120 | FASLG | 13789 | 7513 | 0.002 | 0.3836 | No | ||
| 121 | CACNA1H | 23077 | 7540 | 0.002 | 0.3822 | No | ||
| 122 | AKT2 | 4365 4366 | 7867 | 0.001 | 0.3645 | No | ||
| 123 | MAP2K1IP1 | 12158 | 7868 | 0.001 | 0.3645 | No | ||
| 124 | CACNG4 | 20176 | 8000 | 0.000 | 0.3574 | No | ||
| 125 | RAP1A | 8467 | 8098 | 0.000 | 0.3521 | No | ||
| 126 | FGF7 | 8965 | 8131 | 0.000 | 0.3504 | No | ||
| 127 | RAC3 | 20561 | 8167 | 0.000 | 0.3485 | No | ||
| 128 | CACNG6 | 7033 | 8185 | -0.000 | 0.3475 | No | ||
| 129 | DUSP9 | 24307 | 8206 | -0.000 | 0.3464 | No | ||
| 130 | PLA2G3 | 20968 | 8602 | -0.001 | 0.3250 | No | ||
| 131 | NR4A1 | 9099 | 8658 | -0.001 | 0.3220 | No | ||
| 132 | CACNG3 | 12029 | 8907 | -0.002 | 0.3085 | No | ||
| 133 | PLA2G2A | 16017 | 9066 | -0.002 | 0.3000 | No | ||
| 134 | PLA2G4A | 13809 | 9332 | -0.003 | 0.2856 | No | ||
| 135 | MAP2K4 | 20405 | 9477 | -0.003 | 0.2778 | No | ||
| 136 | MAP2K7 | 6453 | 9567 | -0.003 | 0.2730 | No | ||
| 137 | CACNG8 | 13498 | 9835 | -0.004 | 0.2585 | No | ||
| 138 | RPS6KA3 | 8490 | 9927 | -0.004 | 0.2536 | No | ||
| 139 | PPP3CA | 1863 5284 | 10074 | -0.004 | 0.2458 | No | ||
| 140 | RPS6KA6 | 12458 | 10187 | -0.005 | 0.2397 | No | ||
| 141 | CACNG5 | 1339 20175 | 10230 | -0.005 | 0.2375 | No | ||
| 142 | FGF12 | 1723 22621 | 10236 | -0.005 | 0.2373 | No | ||
| 143 | EVI1 | 4689 8926 | 10259 | -0.005 | 0.2362 | No | ||
| 144 | IL1A | 4915 | 10423 | -0.005 | 0.2274 | No | ||
| 145 | NFATC4 | 22002 | 10475 | -0.006 | 0.2247 | No | ||
| 146 | NFATC2 | 5168 2866 | 10494 | -0.006 | 0.2238 | No | ||
| 147 | MOS | 16254 | 10557 | -0.006 | 0.2205 | No | ||
| 148 | NTF5 | 18248 | 10797 | -0.006 | 0.2076 | No | ||
| 149 | FLNC | 4302 8510 | 10875 | -0.007 | 0.2036 | No | ||
| 150 | BDNF | 14926 2797 | 10903 | -0.007 | 0.2022 | No | ||
| 151 | PDGFRA | 16824 | 10973 | -0.007 | 0.1985 | No | ||
| 152 | MAPK8IP3 | 11402 23080 | 11006 | -0.007 | 0.1969 | No | ||
| 153 | EGFR | 1329 20944 | 11167 | -0.008 | 0.1883 | No | ||
| 154 | FGF11 | 20383 | 11245 | -0.008 | 0.1842 | No | ||
| 155 | PLA2G2E | 16016 | 11290 | -0.008 | 0.1820 | No | ||
| 156 | FGF14 | 21716 3005 3463 | 11291 | -0.008 | 0.1821 | No | ||
| 157 | FGFR1 | 3789 8968 | 11379 | -0.008 | 0.1775 | No | ||
| 158 | MAPKAPK5 | 16387 | 11527 | -0.009 | 0.1696 | No | ||
| 159 | CACNA1B | 8673 4462 | 11626 | -0.009 | 0.1644 | No | ||
| 160 | MAPK10 | 11169 | 11763 | -0.010 | 0.1572 | No | ||
| 161 | RPS6KA2 | 9759 9758 | 11842 | -0.010 | 0.1531 | No | ||
| 162 | CACNB3 | 22373 8679 | 11877 | -0.011 | 0.1514 | No | ||
| 163 | MRAS | 9409 | 11948 | -0.011 | 0.1478 | No | ||
| 164 | RPS6KA5 | 2076 21005 | 12243 | -0.012 | 0.1320 | No | ||
| 165 | PTPRR | 3436 19876 | 12264 | -0.013 | 0.1311 | No | ||
| 166 | FGF5 | 16785 | 12288 | -0.013 | 0.1300 | No | ||
| 167 | CACNA1D | 4464 8675 | 12388 | -0.013 | 0.1248 | No | ||
| 168 | PPP3R2 | 9612 | 12427 | -0.013 | 0.1230 | No | ||
| 169 | FGF20 | 18892 | 12484 | -0.014 | 0.1201 | No | ||
| 170 | CACNA1A | 4461 | 12498 | -0.014 | 0.1196 | No | ||
| 171 | NTRK1 | 15299 | 12524 | -0.014 | 0.1185 | No | ||
| 172 | NGFB | 15476 | 12701 | -0.015 | 0.1092 | No | ||
| 173 | FGF8 | 23656 | 12742 | -0.016 | 0.1072 | No | ||
| 174 | PLA2G2D | 16018 | 12804 | -0.016 | 0.1041 | No | ||
| 175 | FGFR3 | 8969 3566 | 12834 | -0.016 | 0.1028 | No | ||
| 176 | NTRK2 | 9491 | 13241 | -0.020 | 0.0810 | No | ||
| 177 | FGF10 | 4719 8962 | 13261 | -0.020 | 0.0803 | No | ||
| 178 | AKT1 | 8568 | 13355 | -0.021 | 0.0755 | No | ||
| 179 | RASGRF1 | 3050 19357 | 13397 | -0.021 | 0.0736 | No | ||
| 180 | GADD45G | 21637 | 13491 | -0.022 | 0.0689 | No | ||
| 181 | TGFB3 | 10161 | 13520 | -0.023 | 0.0677 | No | ||
| 182 | DAXX | 23290 | 13619 | -0.024 | 0.0628 | No | ||
| 183 | FGF2 | 15608 | 13664 | -0.025 | 0.0608 | No | ||
| 184 | NTF3 | 16991 | 13671 | -0.025 | 0.0608 | No | ||
| 185 | IKBKG | 2570 2562 4908 | 13789 | -0.027 | 0.0548 | No | ||
| 186 | CACNA1C | 1158 1166 24464 4463 8674 | 13840 | -0.028 | 0.0525 | No | ||
| 187 | DUSP14 | 12124 | 14289 | -0.036 | 0.0287 | No | ||
| 188 | PTPN5 | 17817 | 14302 | -0.037 | 0.0286 | No | ||
| 189 | CACNA2D3 | 8677 | 14394 | -0.039 | 0.0243 | No | ||
| 190 | MAPK8IP2 | 77 | 14498 | -0.042 | 0.0193 | No | ||
| 191 | MAPK11 | 2264 9618 22163 | 14587 | -0.045 | 0.0152 | No | ||
| 192 | PLA2G6 | 22209 | 14608 | -0.045 | 0.0148 | No | ||
| 193 | FGF23 | 17275 | 14683 | -0.047 | 0.0115 | No | ||
| 194 | FGF17 | 21757 | 14703 | -0.048 | 0.0112 | No | ||
| 195 | TNFRSF1A | 1181 10206 | 14806 | -0.052 | 0.0064 | No | ||
| 196 | ATF2 | 4418 2759 | 14826 | -0.053 | 0.0062 | No | ||
| 197 | MAP3K2 | 11165 | 14854 | -0.054 | 0.0055 | No | ||
| 198 | RASGRF2 | 21397 | 14906 | -0.056 | 0.0036 | No | ||
| 199 | IL1R1 | 14264 3987 | 14939 | -0.057 | 0.0028 | No | ||
| 200 | RAF1 | 17035 | 15064 | -0.065 | -0.0030 | No | ||
| 201 | TGFB2 | 5743 | 15106 | -0.067 | -0.0042 | No | ||
| 202 | DUSP3 | 1358 13022 | 15107 | -0.067 | -0.0032 | No | ||
| 203 | RASA2 | 4329 | 15110 | -0.068 | -0.0023 | No | ||
| 204 | MAX | 21034 | 15233 | -0.075 | -0.0078 | No | ||
| 205 | MAP2K6 | 20614 1414 | 15246 | -0.076 | -0.0073 | No | ||
| 206 | CACNG1 | 20177 | 15255 | -0.077 | -0.0065 | No | ||
| 207 | RPS6KA1 | 15725 | 15263 | -0.077 | -0.0057 | No | ||
| 208 | MAP3K7 | 16255 | 15267 | -0.077 | -0.0047 | No | ||
| 209 | PAK2 | 10310 5875 | 15410 | -0.087 | -0.0111 | No | ||
| 210 | SRF | 22961 1597 | 15416 | -0.087 | -0.0101 | No | ||
| 211 | PLA2G2F | 15700 | 15466 | -0.091 | -0.0114 | No | ||
| 212 | CACNB1 | 1443 1304 | 15529 | -0.097 | -0.0133 | No | ||
| 213 | FGF3 | 17997 | 15532 | -0.097 | -0.0119 | No | ||
| 214 | STMN1 | 9261 | 15569 | -0.101 | -0.0123 | No | ||
| 215 | MAP3K12 | 6454 11164 | 15806 | -0.129 | -0.0232 | No | ||
| 216 | MAPK8 | 6459 | 15826 | -0.132 | -0.0222 | No | ||
| 217 | MAP2K1 | 19082 | 15878 | -0.140 | -0.0229 | No | ||
| 218 | CACNA1S | 14111 | 16028 | -0.163 | -0.0285 | No | ||
| 219 | HRAS | 4868 | 16140 | -0.184 | -0.0318 | No | ||
| 220 | RASGRP3 | 10767 | 16207 | -0.196 | -0.0324 | No | ||
| 221 | NRAS | 5191 | 16367 | -0.233 | -0.0375 | No | ||
| 222 | MAP2K2 | 19933 | 16484 | -0.264 | -0.0398 | No | ||
| 223 | ATF4 | 22417 2239 | 16600 | -0.293 | -0.0416 | No | ||
| 224 | MKNK1 | 2504 | 16601 | -0.293 | -0.0371 | No | ||
| 225 | MAPKAPK3 | 19003 | 16817 | -0.350 | -0.0435 | No | ||
| 226 | NLK | 5179 5178 | 16855 | -0.362 | -0.0400 | No | ||
| 227 | TP53 | 20822 | 16897 | -0.375 | -0.0365 | No | ||
| 228 | PPP5C | 17952 | 16955 | -0.393 | -0.0337 | No | ||
| 229 | FLNB | 13742 | 17243 | -0.502 | -0.0416 | No | ||
| 230 | IKBKB | 4907 | 17370 | -0.562 | -0.0399 | No | ||
| 231 | FLNA | 24129 | 17448 | -0.596 | -0.0351 | No | ||
| 232 | GADD45B | 19935 | 17459 | -0.605 | -0.0264 | No | ||
| 233 | MAPK12 | 2182 2215 22164 | 17503 | -0.632 | -0.0192 | No | ||
| 234 | FGF13 | 4720 8963 2627 | 17551 | -0.656 | -0.0117 | No | ||
| 235 | CHUK | 23665 | 17636 | -0.706 | -0.0056 | No | ||
| 236 | MAP4K1 | 18313 | 18098 | -1.095 | -0.0140 | No | ||
| 237 | ECSIT | 11248 | 18238 | -1.322 | -0.0015 | No | ||
| 238 | PLA2G12A | 1782 7281 1839 | 18298 | -1.446 | 0.0173 | No |