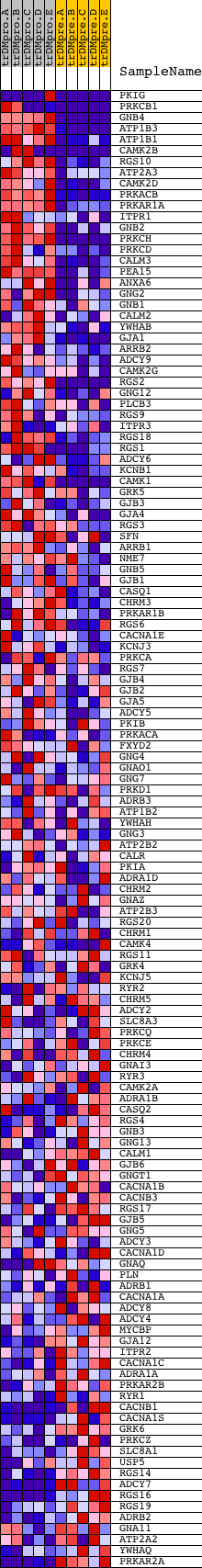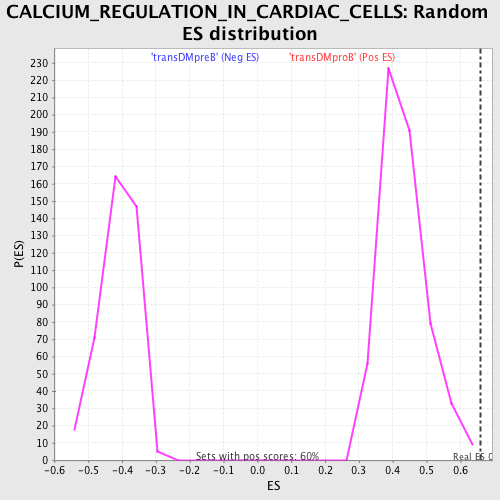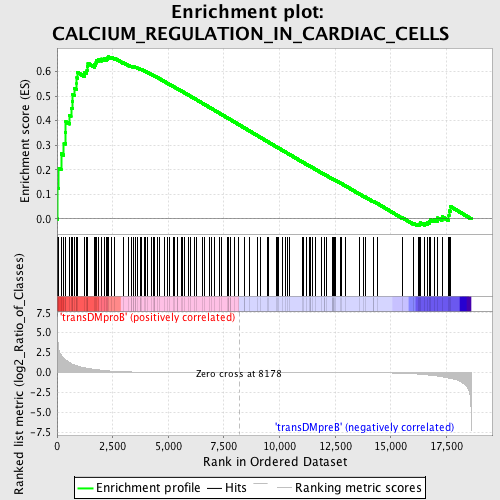
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | CALCIUM_REGULATION_IN_CARDIAC_CELLS |
| Enrichment Score (ES) | 0.6595391 |
| Normalized Enrichment Score (NES) | 1.5251986 |
| Nominal p-value | 0.0016806723 |
| FDR q-value | 0.22204728 |
| FWER p-Value | 0.74 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PKIG | 9577 655 | 22 | 4.334 | 0.1253 | Yes | ||
| 2 | PRKCB1 | 1693 9574 | 81 | 2.858 | 0.2056 | Yes | ||
| 3 | GNB4 | 4787 9028 | 181 | 2.213 | 0.2649 | Yes | ||
| 4 | ATP1B3 | 19037 | 308 | 1.717 | 0.3082 | Yes | ||
| 5 | ATP1B1 | 4420 | 374 | 1.576 | 0.3507 | Yes | ||
| 6 | CAMK2B | 20536 | 391 | 1.552 | 0.3951 | Yes | ||
| 7 | RGS10 | 12591 | 556 | 1.231 | 0.4222 | Yes | ||
| 8 | ATP2A3 | 20795 1314 | 626 | 1.111 | 0.4509 | Yes | ||
| 9 | CAMK2D | 4232 | 680 | 1.045 | 0.4785 | Yes | ||
| 10 | PRKACB | 15140 | 705 | 1.014 | 0.5068 | Yes | ||
| 11 | PRKAR1A | 20615 | 770 | 0.949 | 0.5311 | Yes | ||
| 12 | ITPR1 | 17341 | 884 | 0.841 | 0.5495 | Yes | ||
| 13 | GNB2 | 9026 | 889 | 0.838 | 0.5737 | Yes | ||
| 14 | PRKCH | 21246 | 909 | 0.818 | 0.5966 | Yes | ||
| 15 | PRKCD | 21897 | 1237 | 0.576 | 0.5957 | Yes | ||
| 16 | CALM3 | 8682 | 1340 | 0.528 | 0.6056 | Yes | ||
| 17 | PEA15 | 13753 | 1352 | 0.523 | 0.6203 | Yes | ||
| 18 | ANXA6 | 20449 | 1386 | 0.509 | 0.6333 | Yes | ||
| 19 | GNG2 | 4790 | 1691 | 0.382 | 0.6280 | Yes | ||
| 20 | GNB1 | 15967 | 1745 | 0.363 | 0.6358 | Yes | ||
| 21 | CALM2 | 8681 | 1771 | 0.355 | 0.6448 | Yes | ||
| 22 | YWHAB | 14744 | 1850 | 0.326 | 0.6501 | Yes | ||
| 23 | GJA1 | 20023 | 1992 | 0.287 | 0.6508 | Yes | ||
| 24 | ARRB2 | 20806 | 2116 | 0.251 | 0.6515 | Yes | ||
| 25 | ADCY9 | 22680 | 2238 | 0.219 | 0.6514 | Yes | ||
| 26 | CAMK2G | 21905 | 2283 | 0.210 | 0.6551 | Yes | ||
| 27 | RGS2 | 5379 | 2312 | 0.203 | 0.6595 | Yes | ||
| 28 | GNG12 | 4789 | 2454 | 0.170 | 0.6569 | No | ||
| 29 | PLCB3 | 23799 | 2591 | 0.147 | 0.6538 | No | ||
| 30 | RGS9 | 20172 | 2983 | 0.092 | 0.6354 | No | ||
| 31 | ITPR3 | 9195 | 3225 | 0.069 | 0.6243 | No | ||
| 32 | RGS18 | 13810 | 3330 | 0.061 | 0.6205 | No | ||
| 33 | RGS1 | 3970 6936 | 3350 | 0.059 | 0.6212 | No | ||
| 34 | ADCY6 | 22139 2283 8551 | 3431 | 0.054 | 0.6184 | No | ||
| 35 | KCNB1 | 14334 | 3438 | 0.054 | 0.6197 | No | ||
| 36 | CAMK1 | 1037 1170 17044 | 3520 | 0.048 | 0.6167 | No | ||
| 37 | GRK5 | 9036 23814 | 3534 | 0.048 | 0.6174 | No | ||
| 38 | GJB3 | 15754 | 3626 | 0.043 | 0.6137 | No | ||
| 39 | GJA4 | 15755 | 3756 | 0.037 | 0.6079 | No | ||
| 40 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 3763 | 0.037 | 0.6086 | No | ||
| 41 | SFN | 7060 12062 | 3774 | 0.037 | 0.6092 | No | ||
| 42 | ARRB1 | 3890 8465 18178 4276 | 3916 | 0.032 | 0.6025 | No | ||
| 43 | NME7 | 926 4101 14070 3986 3946 931 | 3991 | 0.029 | 0.5993 | No | ||
| 44 | GNB5 | 2985 9029 3153 | 4045 | 0.028 | 0.5972 | No | ||
| 45 | GJB1 | 24276 | 4228 | 0.022 | 0.5880 | No | ||
| 46 | CASQ1 | 13752 8695 | 4334 | 0.020 | 0.5830 | No | ||
| 47 | CHRM3 | 21547 | 4376 | 0.019 | 0.5813 | No | ||
| 48 | PRKAR1B | 16320 | 4389 | 0.019 | 0.5812 | No | ||
| 49 | RGS6 | 21219 | 4526 | 0.017 | 0.5743 | No | ||
| 50 | CACNA1E | 4465 | 4619 | 0.016 | 0.5698 | No | ||
| 51 | KCNJ3 | 4944 | 4812 | 0.013 | 0.5598 | No | ||
| 52 | PRKCA | 20174 | 4949 | 0.012 | 0.5528 | No | ||
| 53 | RGS7 | 13743 | 5042 | 0.011 | 0.5482 | No | ||
| 54 | GJB4 | 4782 | 5215 | 0.010 | 0.5392 | No | ||
| 55 | GJB2 | 9018 21806 | 5291 | 0.010 | 0.5354 | No | ||
| 56 | GJA5 | 1929 9017 4779 | 5402 | 0.009 | 0.5297 | No | ||
| 57 | ADCY5 | 10312 | 5405 | 0.009 | 0.5299 | No | ||
| 58 | PKIB | 20022 | 5569 | 0.008 | 0.5213 | No | ||
| 59 | PRKACA | 18549 3844 | 5640 | 0.008 | 0.5177 | No | ||
| 60 | FXYD2 | 3038 19470 3068 3039 | 5717 | 0.007 | 0.5138 | No | ||
| 61 | GNG4 | 21706 | 5887 | 0.007 | 0.5049 | No | ||
| 62 | GNAO1 | 4785 3829 | 6003 | 0.006 | 0.4989 | No | ||
| 63 | GNG7 | 19684 | 6195 | 0.006 | 0.4887 | No | ||
| 64 | PRKD1 | 2074 21075 | 6271 | 0.005 | 0.4848 | No | ||
| 65 | ADRB3 | 18901 | 6525 | 0.004 | 0.4712 | No | ||
| 66 | ATP1B2 | 20390 | 6637 | 0.004 | 0.4653 | No | ||
| 67 | YWHAH | 5937 10368 | 6860 | 0.003 | 0.4534 | No | ||
| 68 | GNG3 | 23752 | 6945 | 0.003 | 0.4490 | No | ||
| 69 | ATP2B2 | 17038 | 7094 | 0.003 | 0.4410 | No | ||
| 70 | CALR | 18814 | 7309 | 0.002 | 0.4295 | No | ||
| 71 | PKIA | 15639 | 7402 | 0.002 | 0.4246 | No | ||
| 72 | ADRA1D | 20737 8561 | 7670 | 0.001 | 0.4102 | No | ||
| 73 | CHRM2 | 10840 | 7692 | 0.001 | 0.4091 | No | ||
| 74 | GNAZ | 71 | 7699 | 0.001 | 0.4088 | No | ||
| 75 | ATP2B3 | 24308 11557 | 7803 | 0.001 | 0.4033 | No | ||
| 76 | RGS20 | 4001 12202 | 7807 | 0.001 | 0.4031 | No | ||
| 77 | CHRM1 | 23943 | 7972 | 0.001 | 0.3943 | No | ||
| 78 | CAMK4 | 4473 | 8169 | 0.000 | 0.3837 | No | ||
| 79 | RGS11 | 23329 | 8422 | -0.001 | 0.3701 | No | ||
| 80 | GRK4 | 3467 16873 | 8438 | -0.001 | 0.3693 | No | ||
| 81 | KCNJ5 | 9209 | 8655 | -0.001 | 0.3576 | No | ||
| 82 | RYR2 | 5403 | 9023 | -0.002 | 0.3378 | No | ||
| 83 | CHRM5 | 10038 | 9123 | -0.002 | 0.3325 | No | ||
| 84 | ADCY2 | 21412 | 9456 | -0.003 | 0.3146 | No | ||
| 85 | SLC8A3 | 2065 4293 | 9523 | -0.003 | 0.3112 | No | ||
| 86 | PRKCQ | 2873 2831 | 9877 | -0.004 | 0.2922 | No | ||
| 87 | PRKCE | 9575 | 9884 | -0.004 | 0.2920 | No | ||
| 88 | CHRM4 | 14945 | 9970 | -0.004 | 0.2875 | No | ||
| 89 | GNAI3 | 15198 | 10126 | -0.005 | 0.2792 | No | ||
| 90 | RYR3 | 14486 | 10261 | -0.005 | 0.2721 | No | ||
| 91 | CAMK2A | 2024 23541 1980 | 10341 | -0.005 | 0.2680 | No | ||
| 92 | ADRA1B | 4354 | 10464 | -0.005 | 0.2616 | No | ||
| 93 | CASQ2 | 15477 | 11013 | -0.007 | 0.2321 | No | ||
| 94 | RGS4 | 9724 | 11090 | -0.007 | 0.2282 | No | ||
| 95 | GNB3 | 9027 | 11201 | -0.008 | 0.2225 | No | ||
| 96 | GNG13 | 23338 | 11336 | -0.008 | 0.2155 | No | ||
| 97 | CALM1 | 21184 | 11378 | -0.008 | 0.2135 | No | ||
| 98 | GJB6 | 21805 | 11402 | -0.009 | 0.2125 | No | ||
| 99 | GNGT1 | 17533 | 11486 | -0.009 | 0.2083 | No | ||
| 100 | CACNA1B | 8673 4462 | 11626 | -0.009 | 0.2011 | No | ||
| 101 | CACNB3 | 22373 8679 | 11877 | -0.011 | 0.1878 | No | ||
| 102 | RGS17 | 7133 | 12006 | -0.011 | 0.1812 | No | ||
| 103 | GJB5 | 15753 | 12026 | -0.011 | 0.1805 | No | ||
| 104 | GNG5 | 9030 15393 483 | 12089 | -0.012 | 0.1775 | No | ||
| 105 | ADCY3 | 21329 894 | 12378 | -0.013 | 0.1623 | No | ||
| 106 | CACNA1D | 4464 8675 | 12388 | -0.013 | 0.1622 | No | ||
| 107 | GNAQ | 4786 23909 3685 | 12410 | -0.013 | 0.1615 | No | ||
| 108 | PLN | 20026 | 12440 | -0.014 | 0.1603 | No | ||
| 109 | ADRB1 | 4357 | 12463 | -0.014 | 0.1595 | No | ||
| 110 | CACNA1A | 4461 | 12498 | -0.014 | 0.1581 | No | ||
| 111 | ADCY8 | 22281 | 12754 | -0.016 | 0.1447 | No | ||
| 112 | ADCY4 | 2980 21818 | 12802 | -0.016 | 0.1427 | No | ||
| 113 | MYCBP | 7093 | 12953 | -0.017 | 0.1351 | No | ||
| 114 | GJA12 | 20433 1323 | 13609 | -0.024 | 0.1003 | No | ||
| 115 | ITPR2 | 9194 | 13771 | -0.026 | 0.0924 | No | ||
| 116 | CACNA1C | 1158 1166 24464 4463 8674 | 13840 | -0.028 | 0.0895 | No | ||
| 117 | ADRA1A | 2850 8560 | 14214 | -0.035 | 0.0703 | No | ||
| 118 | PRKAR2B | 5288 2107 | 14236 | -0.035 | 0.0702 | No | ||
| 119 | RYR1 | 17903 17902 9770 | 14405 | -0.039 | 0.0623 | No | ||
| 120 | CACNB1 | 1443 1304 | 15529 | -0.097 | 0.0044 | No | ||
| 121 | CACNA1S | 14111 | 16028 | -0.163 | -0.0178 | No | ||
| 122 | GRK6 | 6449 | 16230 | -0.202 | -0.0228 | No | ||
| 123 | PRKCZ | 5260 | 16309 | -0.219 | -0.0206 | No | ||
| 124 | SLC8A1 | 9830 | 16323 | -0.223 | -0.0148 | No | ||
| 125 | USP5 | 17004 | 16525 | -0.273 | -0.0177 | No | ||
| 126 | RGS14 | 21630 | 16651 | -0.309 | -0.0155 | No | ||
| 127 | ADCY7 | 8552 448 | 16725 | -0.325 | -0.0099 | No | ||
| 128 | RGS16 | 14094 | 16770 | -0.339 | -0.0024 | No | ||
| 129 | RGS19 | 14303 | 16956 | -0.393 | -0.0010 | No | ||
| 130 | ADRB2 | 23422 | 17107 | -0.447 | 0.0040 | No | ||
| 131 | GNA11 | 3300 3345 19670 | 17302 | -0.531 | 0.0090 | No | ||
| 132 | ATP2A2 | 4421 3481 | 17610 | -0.694 | 0.0126 | No | ||
| 133 | YWHAQ | 10369 | 17620 | -0.697 | 0.0325 | No | ||
| 134 | PRKAR2A | 5287 | 17679 | -0.732 | 0.0507 | No |