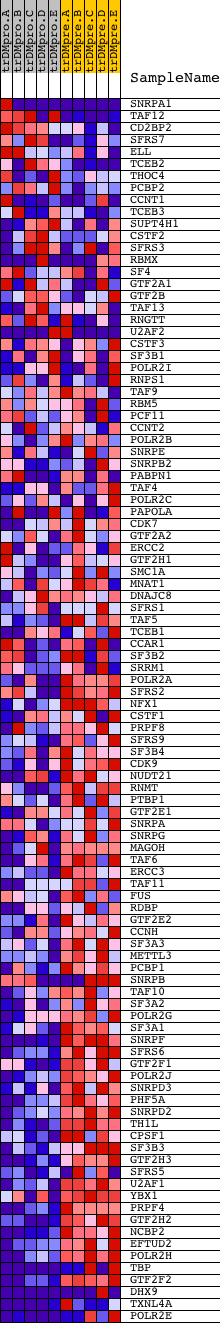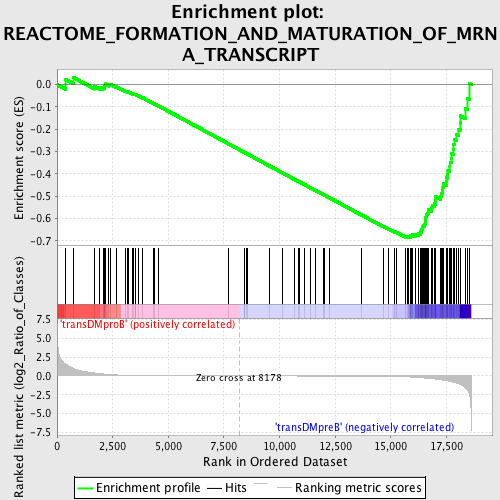
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | -0.6850273 |
| Normalized Enrichment Score (NES) | -1.5862722 |
| Nominal p-value | 0.002202643 |
| FDR q-value | 0.04701631 |
| FWER p-Value | 0.525 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SNRPA1 | 1373 18216 | 381 | 1.570 | 0.0235 | No | ||
| 2 | TAF12 | 2537 16058 2338 | 740 | 0.975 | 0.0316 | No | ||
| 3 | CD2BP2 | 17621 | 1689 | 0.383 | -0.0089 | No | ||
| 4 | SFRS7 | 22889 | 1898 | 0.314 | -0.0113 | No | ||
| 5 | ELL | 18586 | 2065 | 0.266 | -0.0128 | No | ||
| 6 | TCEB2 | 12563 23102 7476 | 2123 | 0.250 | -0.0089 | No | ||
| 7 | THOC4 | 5710 10114 | 2148 | 0.243 | -0.0033 | No | ||
| 8 | PCBP2 | 9533 | 2156 | 0.241 | 0.0031 | No | ||
| 9 | CCNT1 | 22140 11607 | 2331 | 0.197 | -0.0008 | No | ||
| 10 | TCEB3 | 15707 | 2401 | 0.181 | 0.0005 | No | ||
| 11 | SUPT4H1 | 9938 | 2655 | 0.137 | -0.0093 | No | ||
| 12 | CSTF2 | 24257 | 3071 | 0.083 | -0.0293 | No | ||
| 13 | SFRS3 | 5428 23312 | 3158 | 0.076 | -0.0318 | No | ||
| 14 | RBMX | 5367 9708 | 3204 | 0.071 | -0.0323 | No | ||
| 15 | SF4 | 18593 | 3404 | 0.056 | -0.0415 | No | ||
| 16 | GTF2A1 | 2064 8187 8188 13512 | 3452 | 0.053 | -0.0425 | No | ||
| 17 | GTF2B | 10489 | 3509 | 0.049 | -0.0442 | No | ||
| 18 | TAF13 | 8288 | 3542 | 0.047 | -0.0446 | No | ||
| 19 | RNGTT | 10769 2354 6272 | 3673 | 0.041 | -0.0504 | No | ||
| 20 | U2AF2 | 5813 5812 5814 | 3857 | 0.034 | -0.0594 | No | ||
| 21 | CSTF3 | 10449 6002 | 4328 | 0.020 | -0.0842 | No | ||
| 22 | SF3B1 | 13954 | 4374 | 0.019 | -0.0861 | No | ||
| 23 | POLR2I | 12839 | 4549 | 0.016 | -0.0950 | No | ||
| 24 | RNPS1 | 9730 23361 | 7696 | 0.001 | -0.2649 | No | ||
| 25 | TAF9 | 3213 8433 | 8410 | -0.001 | -0.3034 | No | ||
| 26 | RBM5 | 13507 3016 | 8413 | -0.001 | -0.3035 | No | ||
| 27 | PCF11 | 121 | 8517 | -0.001 | -0.3090 | No | ||
| 28 | CCNT2 | 3993 13079 | 8564 | -0.001 | -0.3115 | No | ||
| 29 | POLR2B | 16817 | 9559 | -0.003 | -0.3651 | No | ||
| 30 | SNRPE | 9843 | 10124 | -0.005 | -0.3954 | No | ||
| 31 | SNRPB2 | 5470 | 10690 | -0.006 | -0.4257 | No | ||
| 32 | PABPN1 | 12018 | 10865 | -0.007 | -0.4350 | No | ||
| 33 | TAF4 | 14319 | 10882 | -0.007 | -0.4356 | No | ||
| 34 | POLR2C | 9750 | 11129 | -0.008 | -0.4487 | No | ||
| 35 | PAPOLA | 5261 9585 | 11405 | -0.009 | -0.4633 | No | ||
| 36 | CDK7 | 21365 | 11610 | -0.009 | -0.4741 | No | ||
| 37 | GTF2A2 | 10654 | 11981 | -0.011 | -0.4937 | No | ||
| 38 | ERCC2 | 1549 18363 3812 | 12004 | -0.011 | -0.4946 | No | ||
| 39 | GTF2H1 | 4069 18236 | 12236 | -0.012 | -0.5067 | No | ||
| 40 | SMC1A | 2572 24225 6279 10780 | 13661 | -0.025 | -0.5830 | No | ||
| 41 | MNAT1 | 9396 2161 | 14660 | -0.047 | -0.6356 | No | ||
| 42 | DNAJC8 | 12712 | 14910 | -0.056 | -0.6474 | No | ||
| 43 | SFRS1 | 8492 | 15175 | -0.071 | -0.6597 | No | ||
| 44 | TAF5 | 23833 5934 3759 | 15238 | -0.076 | -0.6609 | No | ||
| 45 | TCEB1 | 13995 | 15676 | -0.113 | -0.6813 | No | ||
| 46 | CCAR1 | 7454 | 15735 | -0.120 | -0.6811 | No | ||
| 47 | SF3B2 | 23974 | 15809 | -0.129 | -0.6814 | Yes | ||
| 48 | SRRM1 | 6958 | 15872 | -0.139 | -0.6808 | Yes | ||
| 49 | POLR2A | 5394 | 15899 | -0.143 | -0.6782 | Yes | ||
| 50 | SFRS2 | 9807 20136 | 15923 | -0.146 | -0.6754 | Yes | ||
| 51 | NFX1 | 2475 | 15957 | -0.151 | -0.6729 | Yes | ||
| 52 | CSTF1 | 12515 | 16090 | -0.175 | -0.6752 | Yes | ||
| 53 | PRPF8 | 20780 1371 | 16105 | -0.178 | -0.6709 | Yes | ||
| 54 | SFRS9 | 16731 | 16235 | -0.203 | -0.6722 | Yes | ||
| 55 | SF3B4 | 22269 | 16255 | -0.207 | -0.6674 | Yes | ||
| 56 | CDK9 | 14619 | 16332 | -0.224 | -0.6652 | Yes | ||
| 57 | NUDT21 | 12665 | 16345 | -0.226 | -0.6595 | Yes | ||
| 58 | RNMT | 7501 | 16372 | -0.234 | -0.6543 | Yes | ||
| 59 | PTBP1 | 5303 | 16386 | -0.238 | -0.6483 | Yes | ||
| 60 | GTF2E1 | 22597 | 16435 | -0.251 | -0.6439 | Yes | ||
| 61 | SNRPA | 7007 11992 7008 | 16440 | -0.252 | -0.6370 | Yes | ||
| 62 | SNRPG | 12622 | 16463 | -0.258 | -0.6310 | Yes | ||
| 63 | MAGOH | 2386 16148 | 16529 | -0.275 | -0.6268 | Yes | ||
| 64 | TAF6 | 16322 891 | 16541 | -0.279 | -0.6195 | Yes | ||
| 65 | ERCC3 | 23605 | 16551 | -0.282 | -0.6121 | Yes | ||
| 66 | TAF11 | 12733 | 16567 | -0.286 | -0.6049 | Yes | ||
| 67 | FUS | 6135 | 16575 | -0.287 | -0.5972 | Yes | ||
| 68 | RDBP | 6589 | 16584 | -0.289 | -0.5895 | Yes | ||
| 69 | GTF2E2 | 18635 | 16599 | -0.293 | -0.5821 | Yes | ||
| 70 | CCNH | 7322 | 16640 | -0.306 | -0.5756 | Yes | ||
| 71 | SF3A3 | 16091 | 16675 | -0.313 | -0.5687 | Yes | ||
| 72 | METTL3 | 21841 | 16680 | -0.314 | -0.5601 | Yes | ||
| 73 | PCBP1 | 17085 | 16841 | -0.359 | -0.5586 | Yes | ||
| 74 | SNRPB | 9842 5469 2736 | 16845 | -0.360 | -0.5487 | Yes | ||
| 75 | TAF10 | 10785 | 16868 | -0.367 | -0.5396 | Yes | ||
| 76 | SF3A2 | 19938 | 16967 | -0.396 | -0.5337 | Yes | ||
| 77 | POLR2G | 23753 | 17011 | -0.414 | -0.5244 | Yes | ||
| 78 | SF3A1 | 7450 | 17019 | -0.417 | -0.5131 | Yes | ||
| 79 | SNRPF | 7645 | 17025 | -0.419 | -0.5016 | Yes | ||
| 80 | SFRS6 | 14751 | 17246 | -0.503 | -0.4993 | Yes | ||
| 81 | GTF2F1 | 13599 | 17263 | -0.510 | -0.4859 | Yes | ||
| 82 | POLR2J | 16672 | 17332 | -0.543 | -0.4743 | Yes | ||
| 83 | SNRPD3 | 12514 | 17340 | -0.547 | -0.4594 | Yes | ||
| 84 | PHF5A | 22194 | 17362 | -0.559 | -0.4448 | Yes | ||
| 85 | SNRPD2 | 8412 | 17505 | -0.632 | -0.4347 | Yes | ||
| 86 | TH1L | 14715 | 17511 | -0.635 | -0.4172 | Yes | ||
| 87 | CPSF1 | 22241 | 17547 | -0.654 | -0.4007 | Yes | ||
| 88 | SF3B3 | 18746 | 17567 | -0.669 | -0.3829 | Yes | ||
| 89 | GTF2H3 | 5551 | 17656 | -0.714 | -0.3676 | Yes | ||
| 90 | SFRS5 | 9808 2062 | 17659 | -0.716 | -0.3476 | Yes | ||
| 91 | U2AF1 | 8431 | 17718 | -0.750 | -0.3297 | Yes | ||
| 92 | YBX1 | 5929 2389 | 17736 | -0.765 | -0.3091 | Yes | ||
| 93 | PRPF4 | 7655 12845 | 17806 | -0.820 | -0.2898 | Yes | ||
| 94 | GTF2H2 | 6236 | 17831 | -0.841 | -0.2675 | Yes | ||
| 95 | NCBP2 | 12643 | 17873 | -0.871 | -0.2453 | Yes | ||
| 96 | EFTUD2 | 1219 20203 | 17956 | -0.931 | -0.2236 | Yes | ||
| 97 | POLR2H | 10888 | 18041 | -1.027 | -0.1993 | Yes | ||
| 98 | TBP | 671 1554 | 18119 | -1.131 | -0.1717 | Yes | ||
| 99 | GTF2F2 | 21750 | 18128 | -1.139 | -0.1401 | Yes | ||
| 100 | DHX9 | 8845 4608 3948 | 18361 | -1.568 | -0.1086 | Yes | ||
| 101 | TXNL4A | 6567 11329 | 18451 | -1.865 | -0.0611 | Yes | ||
| 102 | POLR2E | 3325 19699 | 18536 | -2.490 | 0.0043 | Yes |